The Zur regulon of Corynebacterium glutamicum ATCC 13032
- PMID: 20055984
- PMCID: PMC2823685
- DOI: 10.1186/1471-2164-11-12
The Zur regulon of Corynebacterium glutamicum ATCC 13032
Abstract
Background: Zinc is considered as an essential element for all living organisms, but it can be toxic at large concentrations. Bacteria therefore tightly regulate zinc metabolism. The Cg2502 protein of Corynebacterium glutamicum was a candidate to control zinc metabolism in this species, since it was classified as metalloregulator of the zinc uptake regulator (Zur) subgroup of the ferric uptake regulator (Fur) family of DNA-binding transcription regulators.
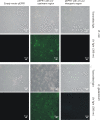
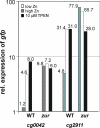
Results: The cg2502 (zur) gene was deleted in the chromosome of C. glutamicum ATCC 13032 by an allelic exchange procedure to generate the zur-deficient mutant C. glutamicum JS2502. Whole-genome DNA microarray hybridizations and real-time RT-PCR assays comparing the gene expression in C. glutamicum JS2502 with that of the wild-type strain detected 18 genes with enhanced expression in the zur mutant. The expression data were combined with results from cross-genome comparisons of shared regulatory sites, revealing the presence of candidate Zur-binding sites in the mapped promoter regions of five transcription units encoding components of potential zinc ABC-type transporters (cg0041-cg0042/cg0043; cg2911-cg2912-cg2913), a putative secreted protein (cg0040), a putative oxidoreductase (cg0795), and a putative P-loop GTPase of the COG0523 protein family (cg0794). Enhanced transcript levels of the respective genes in C. glutamicum JS2502 were verified by real-time RT-PCR, and complementation of the mutant with a wild-type zur gene reversed the effect of differential gene expression. The zinc-dependent expression of the putative cg0042 and cg2911 operons was detected in vivo with a gfp reporter system. Moreover, the zinc-dependent binding of purified Zur protein to double-stranded 40-mer oligonucleotides containing candidate Zur-binding sites was demonstrated in vitro by DNA band shift assays.
Conclusion: Whole-genome expression profiling and DNA band shift assays demonstrated that Zur directly represses in a zinc-dependent manner the expression of nine genes organized in five transcription units. Accordingly, the Zur (Cg2502) protein is the key transcription regulator for genes involved in zinc homeostasis in C. glutamicum.
Figures





References
-
- Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L. et al.The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/S0168-1656(03)00154-8. - DOI - PubMed
-
- Brune I, Brinkrolf K, Kalinowski J, Pühler A, Tauch A. The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences. BMC Genomics. 2005;6:86. doi: 10.1186/1471-2164-6-86. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

