SKPDB: a structural database of shikimate pathway enzymes
- PMID: 20055992
- PMCID: PMC2824673
- DOI: 10.1186/1471-2105-11-12
SKPDB: a structural database of shikimate pathway enzymes
Abstract
Background: The functional and structural characterisation of enzymes that belong to microbial metabolic pathways is very important for structure-based drug design. The main interest in studying shikimate pathway enzymes involves the fact that they are essential for bacteria but do not occur in humans, making them selective targets for design of drugs that do not directly impact humans.
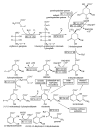
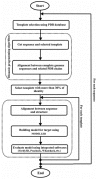
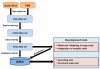
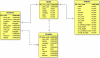
Description: The ShiKimate Pathway DataBase (SKPDB) is a relational database applied to the study of shikimate pathway enzymes in microorganisms and plants. The current database is updated regularly with the addition of new data; there are currently 8902 enzymes of the shikimate pathway from different sources. The database contains extensive information on each enzyme, including detailed descriptions about sequence, references, and structural and functional studies. All files (primary sequence, atomic coordinates and quality scores) are available for downloading. The modeled structures can be viewed using the Jmol program.
Conclusions: The SKPDB provides a large number of structural models to be used in docking simulations, virtual screening initiatives and drug design. It is freely accessible at http://lsbzix.rc.unesp.br/skpdb/.
Figures




References
-
- Jacobson M, Sali A. Comparative Protein Structure Modeling and its Applications to Drug Discovery. Annu Rep Medic Chem. 2004;39:259–276. full_text.
-
- Roberts CW, Roberts F, Lyons RE, Kirisits MJ, Mui EJ, Finnerty J, Johnson JJ, Ferguson DJ, Coggins JR, Krell T, Coombs GH, Milhous WK, Kyle DE, Tzipori S, Barnwell J, Dame JB, Carlton J, McLeod R. The shikimate pathway and its branches in apicomplexan parasites. J Infect Dis. 2002;185:S25–S36. doi: 10.1086/338004. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

