Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer
- PMID: 20056007
- PMCID: PMC2880421
- DOI: 10.1186/bcr2466
Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer
Abstract
Introduction: Ductal carcinoma in situ (DCIS) is a non-invasive lesion of the breast that is frequently detected by mammography and subsequently removed by surgery. However, it is estimated that about half of the detected lesions would never have progressed into invasive cancer. Identifying DCIS and invasive cancer specific epigenetic lesions and understanding how these epigenetic changes are involved in triggering tumour progression is important for a better understanding of which lesions are at risk of becoming invasive.
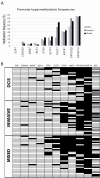
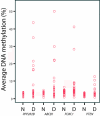
Methods: Quantitative DNA methylation analysis of ABCB1, CDKN2A/p16INK4a, ESR1, FOXC1, GSTP1, IGF2, MGMT, MLH1, PPP2R2B, PTEN and RASSF1A was performed by pyrosequencing in a series of 27 pure DCIS, 28 small invasive ductal carcinomas (IDCs), 34 IDCs with a DCIS component and 5 normal breast tissue samples. FOXC1, ABCB1, PPP2R2B and PTEN were analyzed in 23 additional normal breast tissue samples. Real-Time PCR expression analysis was performed for FOXC1.
Results: Aberrant DNA methylation was observed in all three diagnosis groups for the following genes: ABCB1, FOXC1, GSTP1, MGMT, MLH1, PPP2R2B, PTEN and RASSF1A. For most of these genes, methylation was already present at the DCIS level with the same frequency as within IDCs. For FOXC1 significant differences in methylation levels were observed between normal breast tissue and invasive tumours (P < 0.001). The average DNA methylation levels were significantly higher in the pure IDCs and IDCs with DCIS compared to pure DCIS (P = 0.007 and P = 0.001, respectively). Real-time PCR analysis of FOXC1 expression from 25 DCIS, 23 IDCs and 28 normal tissue samples showed lower gene expression levels of FOXC1 in both methylated and unmethylated tumours compared to normal tissue (P < 0.001). DNA methylation levels of FOXC1, GSTP1, ABCB1 and RASSF1A were higher in oestrogen receptor (ER) positive vs. ER negative tumours; whereas methylation levels of FOXC1, ABCB1, PPP2R2B and PTEN were lower in tumours with a TP53 mutation.
Conclusions: Quantitative methylation analysis identified ABCB1, FOXC1, PPP2R2B and PTEN as novel genes to be methylated in DCIS. In particular, FOXC1 showed a significant increase in the methylation frequency in invasive tumours. Low FOXC1 gene expression in both methylated and unmethylated DCIS and IDCs indicates that the loss of its expression is an early event during breast cancer progression.
Figures


Similar articles
-
Quantitative DNA methylation analyses reveal stage dependent DNA methylation and association to clinico-pathological factors in breast tumors.BMC Cancer. 2013 Oct 5;13:456. doi: 10.1186/1471-2407-13-456. BMC Cancer. 2013. PMID: 24093668 Free PMC article.
-
Promoter CpG island hypermethylation during breast cancer progression.Virchows Arch. 2011 Jan;458(1):73-84. doi: 10.1007/s00428-010-1013-6. Epub 2010 Dec 1. Virchows Arch. 2011. PMID: 21120523
-
DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response.Mol Cancer. 2010 Mar 25;9:68. doi: 10.1186/1476-4598-9-68. Mol Cancer. 2010. PMID: 20338046 Free PMC article.
-
The association between phosphatase and tensin homolog hypermethylation and patients with breast cancer, a meta-analysis and literature review.Sci Rep. 2016 Sep 13;6:32723. doi: 10.1038/srep32723. Sci Rep. 2016. PMID: 27620353 Free PMC article. Review.
-
Carcinoma in situ of the female breast. A clinico-pathological, immunohistological, and DNA ploidy study.APMIS Suppl. 2003;(108):1-67. APMIS Suppl. 2003. PMID: 12874968 Review.
Cited by
-
The association of PTEN hypermethylation and breast cancer: a meta-analysis.Onco Targets Ther. 2016 Sep 12;9:5643-50. doi: 10.2147/OTT.S111684. eCollection 2016. Onco Targets Ther. 2016. PMID: 27672335 Free PMC article.
-
Active and secondhand smoke exposure throughout life and DNA methylation in breast tumors.Cancer Causes Control. 2019 Jan;30(1):53-62. doi: 10.1007/s10552-018-1102-4. Epub 2019 Jan 7. Cancer Causes Control. 2019. PMID: 30617699 Free PMC article.
-
Ductal Carcinoma in Situ Biomarkers in a Precision Medicine Era: Current and Future Molecular-Based Testing.Am J Pathol. 2019 May;189(5):956-965. doi: 10.1016/j.ajpath.2018.08.020. Epub 2018 Oct 29. Am J Pathol. 2019. PMID: 30385093 Free PMC article. Review.
-
FOXC1: an emerging marker and therapeutic target for cancer.Oncogene. 2017 Jul 13;36(28):3957-3963. doi: 10.1038/onc.2017.48. Epub 2017 Mar 13. Oncogene. 2017. PMID: 28288141 Free PMC article. Review.
-
The forkhead box C1 (FOXC1) transcription factor is downregulated in acute promyelocytic leukemia.Oncotarget. 2017 Sep 20;8(48):84074-84085. doi: 10.18632/oncotarget.21101. eCollection 2017 Oct 13. Oncotarget. 2017. PMID: 29137406 Free PMC article.
References
-
- Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. - DOI - PMC - PubMed
-
- Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, Fehm T, Solomayer E, Riess O, Wallwiener D, Kurek R, Neubauer HJ. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66:5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

