Consequences of the pathogenic T9176C mutation of human mitochondrial DNA on yeast mitochondrial ATP synthase
- PMID: 20056103
- PMCID: PMC2891117
- DOI: 10.1016/j.bbabio.2009.12.022
Consequences of the pathogenic T9176C mutation of human mitochondrial DNA on yeast mitochondrial ATP synthase
Abstract
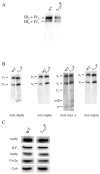
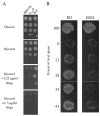
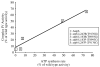
Several human neurological disorders have been associated with various mutations affecting mitochondrial enzymes involved in cellular ATP production. One of these mutations, T9176C in the mitochondrial DNA (mtDNA), changes a highly conserved leucine residue into proline at position 217 of the mitochondrially encoded Atp6p (or a) subunit of the F1FO-ATP synthase. The consequences of this mutation on the mitochondrial ATP synthase are still poorly defined. To gain insight into the primary pathogenic mechanisms induced by T9176C, we have investigated the consequences of this mutation on the ATP synthase of yeast where Atp6p is also encoded by the mtDNA. In vitro, yeast atp6-T9176C mitochondria showed a 30% decrease in the rate of ATP synthesis. When forcing the F1FO complex to work in the reverse mode, i.e. F1-catalyzed hydrolysis of ATP coupled to proton transport out of the mitochondrial matrix, the mutant showed a normal proton-pumping activity and this activity was fully sensitive to oligomycin, an inhibitor of the ATP synthase proton channel. However, under conditions of maximal ATP hydrolytic activity, using non-osmotically protected mitochondria, the mutant ATPase activity was less efficiently inhibited by oligomycin (60% inhibition versus 85% for the wild type control). Blue Native Polyacrylamide Gel Electrophoresis analyses revealed that atp6-T9176C yeast accumulated rather good levels of fully assembled ATP synthase complexes. However, a number of sub-complexes (F1, Atp9p-ring, unassembled alpha-F1 subunits) could be detected as well, presumably because of a decreased stability of Atp6p within the ATP synthase. Although the oxidative phosphorylation capacity was reduced in atp6-T9176C yeast, the number of ATP molecules synthesized per electron transferred to oxygen was similar compared with wild type yeast. It can therefore be inferred that the coupling efficiency within the ATP synthase was mostly unaffected and that the T9176C mutation did not increase the proton permeability of the mitochondrial inner membrane.
Copyright © 2009 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Biochemical consequences in yeast of the human mitochondrial DNA 8993T>C mutation in the ATPase6 gene found in NARP/MILS patients.Biochim Biophys Acta. 2009 May;1793(5):817-24. doi: 10.1016/j.bbamcr.2009.02.011. Epub 2009 Mar 6. Biochim Biophys Acta. 2009. PMID: 19269308
-
A yeast model of the neurogenic ataxia retinitis pigmentosa (NARP) T8993G mutation in the mitochondrial ATP synthase-6 gene.J Biol Chem. 2007 Nov 23;282(47):34039-47. doi: 10.1074/jbc.M703053200. Epub 2007 Sep 12. J Biol Chem. 2007. PMID: 17855363
-
Modulation at a distance of proton conductance through the Saccharomyces cerevisiae mitochondrial F1F0-ATP synthase by variants of the oligomycin sensitivity-conferring protein containing substitutions near the C-terminus.J Bioenerg Biomembr. 2000 Dec;32(6):595-607. doi: 10.1023/a:1005674628249. J Bioenerg Biomembr. 2000. PMID: 15254373
-
[Molecular bases of diseases caused by mutations in genes encoding subunits of ATP synthase].Postepy Biochem. 2018 Dec 29;64(4):304-317. doi: 10.18388/pb.2018_144. Postepy Biochem. 2018. PMID: 30656915 Review. Polish.
-
The oligomycin axis of mitochondrial ATP synthase: OSCP and the proton channel.J Bioenerg Biomembr. 2000 Oct;32(5):507-15. doi: 10.1023/a:1005621125812. J Bioenerg Biomembr. 2000. PMID: 15254386 Review.
Cited by
-
Drug Drop Test: How to Quickly Identify Potential Therapeutic Compounds for Mitochondrial Diseases Using Yeast Saccharomyces cerevisiae.Int J Mol Sci. 2023 Jun 27;24(13):10696. doi: 10.3390/ijms241310696. Int J Mol Sci. 2023. PMID: 37445873 Free PMC article. Review.
-
Probing the pathogenicity of patient-derived variants of MT-ATP6 in yeast.Dis Model Mech. 2023 Apr 1;16(4):dmm049783. doi: 10.1242/dmm.049783. Epub 2023 Apr 21. Dis Model Mech. 2023. PMID: 37083953 Free PMC article.
-
Analysis of MT-ATP8 gene variants reported in patients by modeling in silico and in yeast model organism.Sci Rep. 2023 Jun 20;13(1):9972. doi: 10.1038/s41598-023-36637-9. Sci Rep. 2023. PMID: 37340059 Free PMC article.
-
Variants in Human ATP Synthase Mitochondrial Genes: Biochemical Dysfunctions, Associated Diseases, and Therapies.Int J Mol Sci. 2024 Feb 13;25(4):2239. doi: 10.3390/ijms25042239. Int J Mol Sci. 2024. PMID: 38396915 Free PMC article. Review.
-
Response to immunotherapy in a patient with adult onset Leigh syndrome and T9176C mtDNA mutation.Mol Genet Metab Rep. 2016 Jul 1;8:28-32. doi: 10.1016/j.ymgmr.2016.06.004. eCollection 2016 Sep. Mol Genet Metab Rep. 2016. PMID: 27408822 Free PMC article.
References
-
- Saraste M. Oxidative phosphorylation at the fin de siecle. Science. 1999;283:1488–1493. - PubMed
-
- Schon EA, Santra S, Pallotti F, Girvin ME. Pathogenesis of primary defects in mitochondrial ATP synthesis. Semin Cell Dev Biol. 2001;12:441–448. - PubMed
-
- Houstek J, Pickova A, Vojtiskova A, Mracek T, Pecina P, Jesina P. Mitochondrial diseases and genetic defects of ATP synthase. Biochim Biophys Acta. 2006;1757:1400–1405. - PubMed
-
- Kucharczyk R, Zick M, Bietenhader M, Rak M, Couplan E, Blondel M, Caubet SD, di Rago JP. Mitochondrial ATP synthase disorders: molecular mechanisms and the quest for curative therapeutic approaches. Biochim Biophys Acta. 2009;1793:186–199. - PubMed
-
- Fillingame RH, Angevine CM, Dmitriev OY. Mechanics of coupling proton movements to c-ring rotation in ATP synthase. FEBS Lett. 2003;555:29–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

