Opening up the "Black Box": metabolic phenotyping and metabolome-wide association studies in epidemiology
- PMID: 20056386
- PMCID: PMC4048926
- DOI: 10.1016/j.jclinepi.2009.10.001
Opening up the "Black Box": metabolic phenotyping and metabolome-wide association studies in epidemiology
Abstract
Background: Metabolic phenotyping of humans allows information to be captured on the interactions between dietary, xenobiotic, other lifestyle and environmental exposures, and genetic variation, which together influence the balance between health and disease risks at both individual and population levels.
Objectives: We describe here the main procedures in large-scale metabolic phenotyping and their application to metabolome-wide association (MWA) studies.
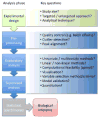
Methods: By use of high-throughput technologies and advanced spectroscopic methods, application of metabolic profiling to large-scale epidemiologic sample collections, including metabolome-wide association (MWA) studies for biomarker discovery and identification.
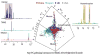
Discussion: Metabolic profiling at epidemiologic scale requires optimization of experimental protocol to maximize reproducibility, sensitivity, and quantitative reliability, and to reduce analytical drift. Customized multivariate statistical modeling approaches are needed for effective data visualization and biomarker discovery with control for false-positive associations since 100s or 1,000s of complex metabolic spectra are being processed.
Conclusion: Metabolic profiling is an exciting addition to the armamentarium of the epidemiologist for the discovery of new disease-risk biomarkers and diagnostics, and to provide novel insights into etiology, biological mechanisms, and pathways.
Conflict of interest statement
Figures
References
-
- Nicholson JK, Wilson I. High resolution proton magnetic resonance spectroscopy of biological fluids. Prog NMR Spect. 1989;21:449–501.
-
- Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–9. - PubMed
-
- Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1(2):153–61. - PubMed
-
- Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455(7216):1054–6. - PubMed
-
- Gavaghan McKee CL, Holmes E, Lenz E, Wilson ID, Nicholson JK. An NMR-based metabonomic approach to investigate the biochemical consequences of genetic strain differences: application to the C57BL10J and Alpk:ApfCD mouse. FEBS Lett. 2000;484(3):169–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

