N-terminal extension of the cholera toxin A1-chain causes rapid degradation after retrotranslocation from endoplasmic reticulum to cytosol
- PMID: 20056601
- PMCID: PMC2825409
- DOI: 10.1074/jbc.M109.062067
N-terminal extension of the cholera toxin A1-chain causes rapid degradation after retrotranslocation from endoplasmic reticulum to cytosol
Abstract
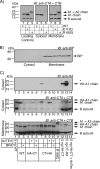
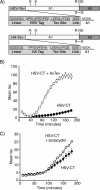
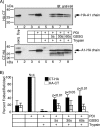
Cholera toxin travels from the plasma membrane to the endoplasmic reticulum of host cells, where a portion of the toxin, the A1-chain, is unfolded and targeted to a protein-conducting channel for retrotranslocation to the cytosol. Unlike most retrotranslocation substrates, the A1-chain escapes degradation by the proteasome and refolds in the cytosol to induce disease. How this occurs remains poorly understood. Here, we show that an unstructured peptide appended to the N terminus of the A1-chain renders the toxin functionally inactive. Cleavage of the peptide extension prior to cell entry rescues toxin half-life and function. The loss of toxicity is explained by rapid degradation by the proteasome after retrotranslocation to the cytosol. Degradation of the mutant toxin does not follow the N-end rule but depends on the two Lys residues at positions 4 and 17 of the native A1-chain, consistent with polyubiquitination at these sites. Thus, retrotranslocation and refolding of the wild-type A1-chain must proceed in a way that protects these Lys residues from attack by E3 ligases.
Figures
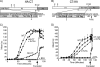


Similar articles
-
Cholera toxin: an intracellular journey into the cytosol by way of the endoplasmic reticulum.Toxins (Basel). 2010 Mar;2(3):310-25. doi: 10.3390/toxins2030310. Epub 2010 Mar 5. Toxins (Basel). 2010. PMID: 22069586 Free PMC article. Review.
-
Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation.EMBO Rep. 2002 Dec;3(12):1222-7. doi: 10.1093/embo-reports/kvf239. Epub 2002 Nov 21. EMBO Rep. 2002. PMID: 12446567 Free PMC article.
-
Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate.J Biol Chem. 2005 Jul 29;280(30):28127-32. doi: 10.1074/jbc.M503138200. Epub 2005 Jun 2. J Biol Chem. 2005. PMID: 15932873
-
Entry of protein toxins into mammalian cells by crossing the endoplasmic reticulum membrane: co-opting basic mechanisms of endoplasmic reticulum-associated degradation.Curr Top Microbiol Immunol. 2005;300:149-68. doi: 10.1007/3-540-28007-3_7. Curr Top Microbiol Immunol. 2005. PMID: 16573240 Review.
-
The ERdj5-Sel1L complex facilitates cholera toxin retrotranslocation.Mol Biol Cell. 2013 Mar;24(6):785-95. doi: 10.1091/mbc.E12-07-0522. Epub 2013 Jan 30. Mol Biol Cell. 2013. PMID: 23363602 Free PMC article.
Cited by
-
A deubiquitinase negatively regulates retro-translocation of nonubiquitinated substrates.Mol Biol Cell. 2013 Nov;24(22):3545-56. doi: 10.1091/mbc.E13-06-0332. Epub 2013 Sep 25. Mol Biol Cell. 2013. PMID: 24068323 Free PMC article.
-
Protein-disulfide isomerase displaces the cholera toxin A1 subunit from the holotoxin without unfolding the A1 subunit.J Biol Chem. 2011 Jun 24;286(25):22090-100. doi: 10.1074/jbc.M111.237966. Epub 2011 May 4. J Biol Chem. 2011. PMID: 21543321 Free PMC article.
-
The cytopathic activity of cholera toxin requires a threshold quantity of cytosolic toxin.Cell Signal. 2023 Jan;101:110520. doi: 10.1016/j.cellsig.2022.110520. Epub 2022 Nov 9. Cell Signal. 2023. PMID: 36371029 Free PMC article.
-
Insights Into the Role of Endoplasmic Reticulum Stress in Infectious Diseases.Front Immunol. 2020 Jan 31;10:3147. doi: 10.3389/fimmu.2019.03147. eCollection 2019. Front Immunol. 2020. PMID: 32082307 Free PMC article. Review.
-
The nucleotide exchange factors Grp170 and Sil1 induce cholera toxin release from BiP to enable retrotranslocation.Mol Biol Cell. 2015 Jun 15;26(12):2181-9. doi: 10.1091/mbc.E15-01-0014. Epub 2015 Apr 15. Mol Biol Cell. 2015. PMID: 25877869 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

