Elevated expression of Paneth cell CRS4C in ileitis-prone SAMP1/YitFc mice: regional distribution, subcellular localization, and mechanism of action
- PMID: 20056603
- PMCID: PMC2844197
- DOI: 10.1074/jbc.M109.083220
Elevated expression of Paneth cell CRS4C in ileitis-prone SAMP1/YitFc mice: regional distribution, subcellular localization, and mechanism of action
Abstract
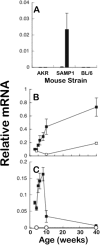
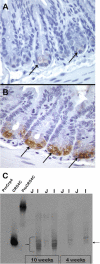
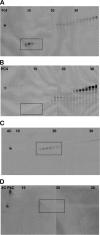
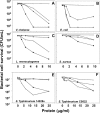
Paneth cells at the base of small intestinal crypts of Lieberkühn secrete host defense peptides and proteins, including alpha-defensins, as mediators of innate immunity. Mouse Paneth cells also express alpha-defensin-related Defcr-rs genes that code for cysteine-rich sequence 4C (CRS4C) peptides that have a unique CPX triplet repeat motif. In ileitis-prone SAMP1/YitFc mice, Paneth cell levels of CRS4C mRNAs and peptides are induced more than a 1000-fold relative to non-prone strains as early as 4 weeks of age, with the mRNA and peptide levels highest in distal ileum and below detection in duodenum. CRS4C-1 peptides are found exclusively in Paneth cells where they occur only in dense core granules and thus are secreted to function in the intestinal lumen. CRS4C bactericidal peptide activity is membrane-disruptive in that it permeabilizes Escherichia coli and induces rapid microbial cell K(+) efflux, but in a manner different from mouse alpha-defensin cryptdin-4. In in vitro studies, inactive pro-CRS4C-1 is converted to bactericidal CRS4C-1 peptide by matrix metalloproteinase-7 (MMP-7) proteolysis of the precursor proregion at the same residue positions that MMP-7 activates mouse pro-alpha-defensins. The absence of processed CRS4C in protein extracts of MMP-7-null mouse ileum demonstrates the in vivo requirement for intracellular MMP-7 in pro-CRS4C processing.
Figures



Similar articles
-
A family of defensin-like genes codes for diverse cysteine-rich peptides in mouse Paneth cells.Genomics. 1994 Nov 1;24(1):99-109. doi: 10.1006/geno.1994.1586. Genomics. 1994. PMID: 7896294
-
Activation of Paneth cell alpha-defensins in mouse small intestine.J Biol Chem. 2002 Feb 15;277(7):5219-28. doi: 10.1074/jbc.M109410200. Epub 2001 Dec 3. J Biol Chem. 2002. PMID: 11733520
-
Regional variations in Paneth cell antimicrobial peptide expression along the mouse intestinal tract.BMC Immunol. 2008 Jul 17;9:37. doi: 10.1186/1471-2172-9-37. BMC Immunol. 2008. PMID: 18637162 Free PMC article.
-
Paneth cell α-defensins in enteric innate immunity.Cell Mol Life Sci. 2011 Jul;68(13):2215-29. doi: 10.1007/s00018-011-0714-6. Epub 2011 May 11. Cell Mol Life Sci. 2011. PMID: 21560070 Free PMC article. Review.
-
Paneth cell defensins: endogenous peptide components of intestinal host defense.FASEB J. 1996 Sep;10(11):1280-9. doi: 10.1096/fasebj.10.11.8836041. FASEB J. 1996. PMID: 8836041 Review.
Cited by
-
A Strong Impact of Genetic Background on Gut Microflora in Mice.Int J Inflam. 2010 Jun 1;2010(2010):986046. doi: 10.4061/2010/986046. Int J Inflam. 2010. PMID: 20976020 Free PMC article.
-
Effect of bifidobacterium on defensin-5 expression in intestinal injury of preweaning rats.World J Gastroenterol. 2015 Mar 7;21(9):2638-44. doi: 10.3748/wjg.v21.i9.2638. World J Gastroenterol. 2015. PMID: 25759531 Free PMC article.
-
Guardians of the Gut: Enteric Defensins.Front Microbiol. 2017 Apr 19;8:647. doi: 10.3389/fmicb.2017.00647. eCollection 2017. Front Microbiol. 2017. PMID: 28469609 Free PMC article. Review.
-
SAMP1/YitFc mouse strain: a spontaneous model of Crohn's disease-like ileitis.Inflamm Bowel Dis. 2011 Dec;17(12):2566-84. doi: 10.1002/ibd.21638. Epub 2011 May 6. Inflamm Bowel Dis. 2011. PMID: 21557393 Free PMC article. Review.
-
A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions.Mucosal Immunol. 2015 Mar;8(2):352-61. doi: 10.1038/mi.2014.72. Epub 2014 Aug 13. Mucosal Immunol. 2015. PMID: 25118165 Free PMC article.
References
-
- Lehrer R. I. (2007) Curr. Opin. Hematol. 14, 16–21 - PubMed
-
- Selsted M. E., Ouellette A. J. (2005) Nat. Immunol. 6, 551–557 - PubMed
-
- Brogden K. A., Ackermann M., McCray P. B., Jr., Tack B. F. (2003) Int. J. Antimicrob. Agents 22, 465–478 - PubMed
-
- Schutte B. C., McCray P. B., Jr. (2002) Annu. Rev. Physiol. 64, 709–748 - PubMed
-
- Tomasinsig L., Zanetti M. (2005) Curr. Protein. Pept. Sci. 6, 23–34 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

