Multi-step control of muscle diversity by Hox proteins in the Drosophila embryo
- PMID: 20056681
- PMCID: PMC2858909
- DOI: 10.1242/dev.045286
Multi-step control of muscle diversity by Hox proteins in the Drosophila embryo
Abstract
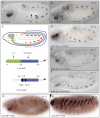
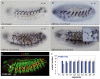
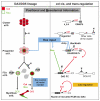
Hox transcription factors control many aspects of animal morphogenetic diversity. The segmental pattern of Drosophila larval muscles shows stereotyped variations along the anteroposterior body axis. Each muscle is seeded by a founder cell and the properties specific to each muscle reflect the expression by each founder cell of a specific combination of 'identity' transcription factors. Founder cells originate from asymmetric division of progenitor cells specified at fixed positions. Using the dorsal DA3 muscle lineage as a paradigm, we show here that Hox proteins play a decisive role in establishing the pattern of Drosophila muscles by controlling the expression of identity transcription factors, such as Nautilus and Collier (Col), at the progenitor stage. High-resolution analysis, using newly designed intron-containing reporter genes to detect primary transcripts, shows that the progenitor stage is the key step at which segment-specific information carried by Hox proteins is superimposed on intrasegmental positional information. Differential control of col transcription by the Antennapedia and Ultrabithorax/Abdominal-A paralogs is mediated by separate cis-regulatory modules (CRMs). Hox proteins also control the segment-specific number of myoblasts allocated to the DA3 muscle. We conclude that Hox proteins both regulate and contribute to the combinatorial code of transcription factors that specify muscle identity and act at several steps during the muscle-specification process to generate muscle diversity.
Figures
Similar articles
-
Requirement for the Drosophila COE transcription factor Collier in formation of an embryonic muscle: transcriptional response to notch signalling.Development. 1999 Apr;126(7):1495-504. doi: 10.1242/dev.126.7.1495. Development. 1999. PMID: 10068642
-
Segmental variations in the patterns of somatic muscles: what roles for Hox?Fly (Austin). 2010 Jul-Sep;4(3):249-52. doi: 10.4161/fly.4.3.12281. Fly (Austin). 2010. PMID: 20519967
-
Collier transcription in a single Drosophila muscle lineage: the combinatorial control of muscle identity.Development. 2007 Dec;134(24):4347-55. doi: 10.1242/dev.008409. Epub 2007 Nov 14. Development. 2007. PMID: 18003742
-
Implications of the spatial and temporal regulation of Hox genes on development and evolution.Int J Dev Biol. 1998;42(3):437-44. Int J Dev Biol. 1998. PMID: 9654029 Review.
-
Cellular analysis of newly identified Hox downstream genes in Drosophila.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):273-8. doi: 10.1016/j.ejcb.2009.11.012. Epub 2009 Dec 16. Eur J Cell Biol. 2010. PMID: 20018403 Review.
Cited by
-
Genetic Control of Muscle Diversification and Homeostasis: Insights from Drosophila.Cells. 2020 Jun 25;9(6):1543. doi: 10.3390/cells9061543. Cells. 2020. PMID: 32630420 Free PMC article. Review.
-
Specification of the somatic musculature in Drosophila.Wiley Interdiscip Rev Dev Biol. 2015 Jul-Aug;4(4):357-75. doi: 10.1002/wdev.182. Epub 2015 Feb 27. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 25728002 Free PMC article. Review.
-
Genome-Wide Analysis in Drosophila Reveals the Genetic Basis of Variation in Age-Specific Physical Performance and Response to ACE Inhibition.Genes (Basel). 2022 Jan 14;13(1):143. doi: 10.3390/genes13010143. Genes (Basel). 2022. PMID: 35052483 Free PMC article.
-
Myoblast fusion: lessons from flies and mice.Development. 2012 Feb;139(4):641-56. doi: 10.1242/dev.068353. Development. 2012. PMID: 22274696 Free PMC article. Review.
-
A machine learning approach for identifying novel cell type-specific transcriptional regulators of myogenesis.PLoS Genet. 2012;8(3):e1002531. doi: 10.1371/journal.pgen.1002531. Epub 2012 Mar 8. PLoS Genet. 2012. PMID: 22412381 Free PMC article.
References
-
- Affolter M., Slattery, M. and Mann, R. S. (2008). A lexicon for homeodomain-DNA recognition. Cell 133, 1133-1135. - PubMed
-
- Alvares L. E., Schubert, F. R., Thorpe, C., Mootoosamy, R. C., Cheng, L., Parkyn, G., Lumsden, A. and Dietrich, S. (2003). Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. Dev. Cell 5, 379-390. - PubMed
-
- Balagopalan L., Keller, C. A. and Abmayr, S. M. (2001). Loss-of-function mutations reveal that the Drosophila nautilus gene is not essential for embryonic myogenesis or viability. Dev. Biol. 231, 374-382. - PubMed
-
- Barolo S., Carver, L. A. and Posakony, J. W. (2000). GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29, 726, 728, 730, 732. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

