Dopamine receptors in a songbird brain
- PMID: 20058221
- PMCID: PMC2904815
- DOI: 10.1002/cne.22255
Dopamine receptors in a songbird brain
Abstract
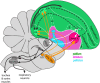
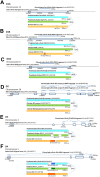
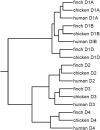
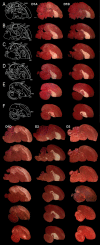
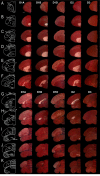
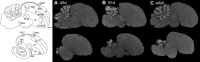
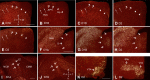
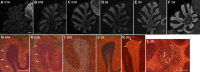
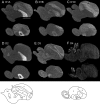
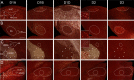
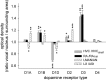
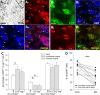
Dopamine is a key neuromodulatory transmitter in the brain. It acts through dopamine receptors to affect changes in neural activity, gene expression, and behavior. In songbirds, dopamine is released into the striatal song nucleus Area X, and the levels depend on social contexts of undirected and directed singing. This differential release is associated with differential expression of activity-dependent genes, such as egr1 (avian zenk), which in mammalian brain are modulated by dopamine receptors. Here we cloned from zebra finch brain cDNAs of all avian dopamine receptors: the D1 (D1A, D1B, D1D) and D2 (D2, D3, D4) families. Comparative sequence analyses of predicted proteins revealed expected phylogenetic relationships, in which the D1 family exists as single exon and the D2 family exists as spliced exon genes. In both zebra finch and chicken, the D1A, D1B, and D2 receptors were highly expressed in the striatum, the D1D and D3 throughout the pallium and within the mesopallium, respectively, and the D4 mainly in the cerebellum. Furthermore, within the zebra finch, all receptors, except for D4, showed differential expression in song nuclei relative to the surrounding regions and developmentally regulated expression that decreased for most receptors during the sensory acquisition and sensorimotor phases of song learning. Within Area X, half of the cells expressed both D1A and D2 receptors, and a higher proportion of the D1A-only-containing neurons expressed egr1 during undirected but not during directed singing. Our findings are consistent with hypotheses that dopamine receptors may be involved in song development and social context-dependent behaviors.
Figures


Similar articles
-
D2 dopamine receptor activation induces female preference for male song in the monogamous zebra finch.J Exp Biol. 2019 Mar 8;222(Pt 5):jeb191510. doi: 10.1242/jeb.191510. J Exp Biol. 2019. PMID: 30850509
-
Differences in dopamine and opioid receptor ratios in the nucleus accumbens relate to physical contact and undirected song in pair-bonded zebra finches.Behav Neurosci. 2022 Feb;136(1):72-83. doi: 10.1037/bne0000494. Epub 2021 Oct 7. Behav Neurosci. 2022. PMID: 34618494 Free PMC article.
-
D1-like dopamine receptor density in nuclei involved in social behavior correlates with song in a context-dependent fashion in male European starlings.Neuroscience. 2009 Mar 31;159(3):962-73. doi: 10.1016/j.neuroscience.2009.01.042. Epub 2009 Jan 27. Neuroscience. 2009. PMID: 19356680 Free PMC article.
-
Dopamine D4-like receptors in vertebrate retina: does the retina offer a model for the D4-receptor analysis?Pol J Pharmacol. 1997 Aug;49(4):201-11. Pol J Pharmacol. 1997. PMID: 9437763 Review.
-
Molecular characteristics of mammalian dopamine receptors.Pharmacol Toxicol. 1997 Sep;81(3):105-13. doi: 10.1111/j.1600-0773.1997.tb00039.x. Pharmacol Toxicol. 1997. PMID: 9335067 Review.
Cited by
-
Context-dependent activation of a social behavior brain network associates with learned vocal production.Res Sq [Preprint]. 2023 Feb 17:rs.3.rs-2587773. doi: 10.21203/rs.3.rs-2587773/v1. Res Sq. 2023. Update in: Brain Struct Funct. 2023 Sep;228(7):1785-1797. doi: 10.1007/s00429-023-02693-0. PMID: 36824963 Free PMC article. Updated. Preprint.
-
The Acute Pharmacological Manipulation of Dopamine Receptors Modulates Judgment Bias in Japanese Quail.Front Physiol. 2022 May 11;13:883021. doi: 10.3389/fphys.2022.883021. eCollection 2022. Front Physiol. 2022. PMID: 35634149 Free PMC article.
-
Individual differences in the motivation to communicate relate to levels of midbrain and striatal catecholamine markers in male European starlings.Horm Behav. 2011 Nov;60(5):529-39. doi: 10.1016/j.yhbeh.2011.08.001. Epub 2011 Aug 31. Horm Behav. 2011. PMID: 21907203 Free PMC article.
-
Dopamine receptor genes and evolutionary differentiation in the domestication of fighting cocks and long-crowing chickens.PLoS One. 2014 Jul 31;9(7):e101778. doi: 10.1371/journal.pone.0101778. eCollection 2014. PLoS One. 2014. PMID: 25078403 Free PMC article.
-
Origins of basal ganglia output signals in singing juvenile birds.J Neurophysiol. 2015 Feb 1;113(3):843-55. doi: 10.1152/jn.00635.2014. Epub 2014 Nov 12. J Neurophysiol. 2015. PMID: 25392171 Free PMC article.
References
-
- Aizman O, Brismar H, Uhlen P, Zettergren E, Levey AI, Forssberg H, Greengard P, Aperia A. Anatomical and physiological evidence for D1 and D2 dopamine receptor colocalization in neostriatal neurons. Nat Neurosci. 2000;3:226–230. - PubMed
-
- Appeltants D, Absil P, Balthazart J, Ball GF. Identification of the origin of catecholaminergic inputs to HVc in canaries by retrograde tract tracing combined with tyrosine hydroxylase immunocytochemistry. J Chem Neuroanat. 2000;18:117–133. - PubMed
-
- Ariano MA, Larson ER, Noblett KL, Sibley DR, Levine MS. Coexpression of striatal dopamine receptor subtypes and excitatory amino acid subunits. Synapse. 1997;26:400–414. - PubMed
-
- Aronov D, Andalman AS, Fee MS. A specialized forebrain circuit for vocal babbling in the juvenile songbird. Science. 2008;320:630–634. - PubMed
-
- Bailey JN, Breidenthal SE, Jorgensen MJ, McCracken JT, Fairbanks LA. The association of DRD4 and novelty seeking is found in a nonhuman primate model. Psychiatr Genet. 2007;17:23–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

