The transcriptome of the fetal inflammatory response syndrome
- PMID: 20059468
- PMCID: PMC3437779
- DOI: 10.1111/j.1600-0897.2009.00791.x
The transcriptome of the fetal inflammatory response syndrome
Abstract
Problem: The fetal inflammatory response syndrome (FIRS) is considered the counterpart of the systemic inflammatory response syndrome (SIRS), but similarities in their regulatory mechanisms are unclear. This study characterizes the fetal mRNA transcriptome of peripheral leukocytes to identify key biological processes and pathways involved in FIRS.
Method of study: Umbilical cord blood from preterm neonates with FIRS (funisitis, plasma IL-6 >11 pg/mL; n = 10) and neonates with no evidence of inflammation (n = 10) was collected at birth. Results Microarray analysis of leukocyte RNA revealed differential expression of 541 unique genes, changes confirmed by qRT-PCR for 41 or 44 genes tested. Similar to SIRS and sepsis, ontological and pathway analyses yielded significant enrichment of biological processes including antigen processing and presentation, immune response, and processes critical to cellular metabolism.
Results: are comparable with microarray studies of endotoxin challenge models and pediatric sepsis, identifying 25 genes across all studies.
Conclusion: This study is the first to profile genome-wide expression in FIRS, which demonstrates a substantial degree of similarity with SIRS despite differences in fetal and adult immune systems.
Figures
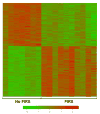
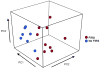




References
-
- Romero R, Gomez R, Ghezzi F, Yoon BH, Mazor M, Edwin SS, Berry SM. A fetal systemic inflammatory response is followed by the spontaneous onset of preterm parturition. Am J Obstet Gynecol. 1998;179:186–193. - PubMed
-
- Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. The fetal inflammatory response syndrome. Am J Obstet Gynecol. 1998;179:194–202. - PubMed
-
- Naeye RL, Ross SM. Amniotic fluid infection syndrome. Clin Obstet Gynaecol. 1982;9:593–607. - PubMed
-
- Minkoff H. Prematurity: infection as an etiologic factor. Obstet Gynecol. 1983;62:137–144. - PubMed
-
- Romero R, Mazor M. Infection and preterm labor. Clin Obstet Gynecol. 1988;31:553–584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

