Establishing a role for the GTPase Ypt1p at the late Golgi
- PMID: 20059749
- PMCID: PMC2861988
- DOI: 10.1111/j.1600-0854.2010.01031.x
Establishing a role for the GTPase Ypt1p at the late Golgi
Abstract
GTPases of the Rab family cycle between an inactive (GDP-bound) and active (GTP-bound) conformation. The active form of the Rab regulates a variety of cellular functions via multiple effectors. Guanine nucleotide exchange factors (GEFs) activate Rabs by accelerating the exchange of GDP for GTP, while GTPase activating proteins (GAPs) inactivate Rabs by stimulating the hydrolysis of GTP. The GTPase Ypt1p is required for endoplasmic reticulum (ER)-Golgi and intra-Golgi traffic in the yeast Saccharomyces cerevisiae. Recent findings, however, have shown that Ypt1p GEF, GAP and an effector are all required for traffic from the early endosome to the Golgi. Here we describe a screen for ypt1 mutants that block traffic from the early endosome to the late Golgi, but not general secretion. This screen has led to the identification of a collection of recessive and dominant mutants that block traffic from the early endosome. While it has long been known that Ypt1p regulates the flow of biosynthetic traffic into the cis side of the Golgi, these findings have established a role for Ypt1p in the regulation of early endosome-Golgi traffic. We propose that Ypt1p regulates the flow of traffic into the cis and trans side of the Golgi via multiple effectors.
Keywords: Golgi; Rab; early endosome; membrane traffic.
Figures




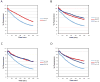
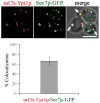



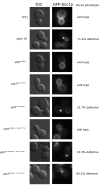
Similar articles
-
Identification of regulators for Ypt1 GTPase nucleotide cycling.Mol Biol Cell. 1998 Oct;9(10):2819-37. doi: 10.1091/mbc.9.10.2819. Mol Biol Cell. 1998. PMID: 9763446 Free PMC article.
-
A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway.Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14408-13. doi: 10.1073/pnas.0906536106. Epub 2009 Aug 4. Proc Natl Acad Sci U S A. 2009. PMID: 19666511 Free PMC article.
-
Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs.J Biol Chem. 2002 Oct 25;277(43):41023-31. doi: 10.1074/jbc.M205783200. Epub 2002 Aug 19. J Biol Chem. 2002. PMID: 12189143
-
[Rab GTPases networks in membrane traffic in Saccharomyces cerevisiae].Yakugaku Zasshi. 2015;135(3):483-92. doi: 10.1248/yakushi.14-00246. Yakugaku Zasshi. 2015. PMID: 25759056 Review. Japanese.
-
Regulation of membrane traffic by Rab GEF and GAP cascades.Small GTPases. 2016 Oct;7(4):252-256. doi: 10.1080/21541248.2016.1213781. Epub 2016 Jul 18. Small GTPases. 2016. PMID: 27427966 Free PMC article. Review.
Cited by
-
α-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding.J Cell Biol. 2011 Jul 11;194(1):89-103. doi: 10.1083/jcb.201011118. J Cell Biol. 2011. PMID: 21746853 Free PMC article.
-
Molecular architecture of the TRAPPII complex and implications for vesicle tethering.Nat Struct Mol Biol. 2010 Nov;17(11):1298-304. doi: 10.1038/nsmb.1914. Epub 2010 Oct 24. Nat Struct Mol Biol. 2010. PMID: 20972447 Free PMC article.
-
A role for endoplasmic reticulum exit sites in foot-and-mouth disease virus infection.J Gen Virol. 2013 Dec;94(Pt 12):2636-2646. doi: 10.1099/vir.0.055442-0. Epub 2013 Aug 20. J Gen Virol. 2013. PMID: 23963534 Free PMC article.
-
Regulation of Golgi Cisternal Progression by Ypt/Rab GTPases.Dev Cell. 2016 Feb 22;36(4):440-52. doi: 10.1016/j.devcel.2016.01.016. Dev Cell. 2016. PMID: 26906739 Free PMC article.
-
Modular TRAPP complexes regulate intracellular protein trafficking through multiple Ypt/Rab GTPases in Saccharomyces cerevisiae.Genetics. 2012 Jun;191(2):451-60. doi: 10.1534/genetics.112.139378. Epub 2012 Mar 16. Genetics. 2012. PMID: 22426882 Free PMC article.
References
-
- Pfeffer SR. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11(12):487–491. - PubMed
-
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–117. - PubMed
-
- Pfeffer SR. Structural clues to Rab GTPase functional diversity. J Biol Chem. 2005;280(16):15485–15488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

