Up- or downregulation of tescalcin in HL-60 cells is associated with their differentiation to either granulocytic or macrophage-like lineage
- PMID: 20060826
- PMCID: PMC2849885
- DOI: 10.1016/j.yexcr.2010.01.007
Up- or downregulation of tescalcin in HL-60 cells is associated with their differentiation to either granulocytic or macrophage-like lineage
Abstract
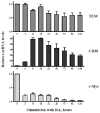
Tescalcin is a 25-kDa EF-hand Ca(2+)-binding protein that is differentially expressed in several mammalian tissues. Previous studies demonstrated that expression of this protein is essential for differentiation of hematopoietic precursor cell lines and primary stem cells into megakaryocytes. Here we show that tescalcin is expressed in primary human granulocytes and is upregulated in human promyelocytic leukemia HL-60 cells that have been induced to differentiate along the granulocytic lineage. However, during induced macrophage-like differentiation of HL-60 cells the expression of tescalcin is downregulated. The decrease in expression is associated with a rapid drop in tescalcin mRNA level, whereas upregulation occurs via a post-transcriptional mechanism. Tescalcin is necessary for HL-60 differentiation into granulocytes as its knockdown by shRNA impairs the ability of HL-60 cells to acquire the characteristic phenotypes such as phagocytic activity and generation of reactive oxygen species measured by respiratory burst assay. Both up- and downregulation of tescalcin require activation of the MEK/ERK cascade. It appears that commitment of HL-60 cells toward granulocytic versus macrophage-like lineage correlates with expression of tescalcin and kinetics of ERK activation. In retinoic acid-induced granulocytic differentiation, the activation of ERK and upregulation of tescalcin occurs slowly (16-48 h). In contrast, in PMA-induced macrophage-like differentiation the activation of ERK is rapid (15-30 min) and tescalcin is downregulated. These studies indicate that tescalcin is one of the key gene products that is involved in switching differentiation program in some cell types.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures









References
-
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33:451–458. - PubMed
-
- Dalton WT, Jr, Ahearn MJ, McCredie KB, Freireich EJ, Stass SA, Trujillo JM. HL-60 cell line was derived from a patient with FAB-M2 and not FAB-M3. Blood. 1988;71:242–247. - PubMed
-
- Collins SJ. The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood. 1987;70:1233–1244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

