The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection
- PMID: 20060839
- PMCID: PMC2824034
- DOI: 10.1016/j.jmb.2009.12.047
The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection
Abstract
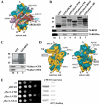
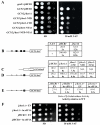
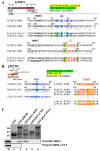
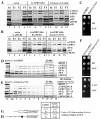
Despite recent progress in our understanding of the numerous functions of individual subunits of eukaryotic translation initiation factor (eIF) 3, little is known on the molecular level. Using NMR spectroscopy, we determined the first solution structure of an interaction between eIF3 subunits. We revealed that a conserved tryptophan residue in the human eIF3j N-terminal acidic motif (NTA) is held in the helix alpha1 and loop 5 hydrophobic pocket of the human eIF3b RNA recognition motif (RRM). Mutating the corresponding "pocket" residues in its yeast orthologue reduces cellular growth rate, eliminates eIF3j/HCR1 association with eIF3b/PRT1 in vitro and in vivo, affects 40S occupancy of eIF3, and produces a leaky scanning defect indicative of a deregulation of the AUG selection process. Unexpectedly, we found that the N-terminal half of eIF3j/HCR1 containing the NTA is indispensable and sufficient for wild-type growth of yeast cells. Furthermore, we demonstrate that deletion of either j/HCR1 or its N-terminal half only, or mutation of the key tryptophan residues results in the severe leaky scanning phenotype partially suppressible by overexpressed eIF1A, which is thought to stabilize properly formed preinitiation complexes at the correct start codon. These findings indicate that eIF3j/HCR1 remains associated with the scanning preinitiation complexes and does not dissociate from the small ribosomal subunit upon mRNA recruitment, as previously believed. Finally, we provide further support for earlier mapping of the ribosomal binding site for human eIF3j by identifying specific interactions of eIF3j/HCR1 with small ribosomal proteins RPS2 and RPS23 located in the vicinity of the mRNA entry channel. Taken together, we propose that eIF3j/HCR1 closely cooperates with the eIF3b/PRT1 RRM and eIF1A on the ribosome to ensure proper formation of the scanning-arrested conformation required for stringent AUG recognition.
(c) 2009 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast.Mol Cell Biol. 2006 Apr;26(8):2984-98. doi: 10.1128/MCB.26.8.2984-2998.2006. Mol Cell Biol. 2006. PMID: 16581774 Free PMC article.
-
The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons.Mol Cell Biol. 2010 Sep;30(18):4415-34. doi: 10.1128/MCB.00280-10. Epub 2010 Jun 28. Mol Cell Biol. 2010. PMID: 20584985 Free PMC article.
-
Crystal structure of the RNA recognition motif of yeast translation initiation factor eIF3b reveals differences to human eIF3b.PLoS One. 2010 Sep 16;5(9):e12784. doi: 10.1371/journal.pone.0012784. PLoS One. 2010. PMID: 20862284 Free PMC article.
-
In vivo deletion analysis of the architecture of a multiprotein complex of translation initiation factors.Methods Enzymol. 2007;431:15-32. doi: 10.1016/S0076-6879(07)31002-1. Methods Enzymol. 2007. PMID: 17923228 Review.
-
Conservation and diversity in the structure of translation initiation factor EIF3 from humans and yeast.Biochimie. 1996;78(11-12):903-7. doi: 10.1016/s0300-9084(97)86711-9. Biochimie. 1996. PMID: 9150866 Review.
Cited by
-
Reprogramming mRNA Expression in Response to Defect in RNA Polymerase III Assembly in the Yeast Saccharomyces cerevisiae.Int J Mol Sci. 2021 Jul 7;22(14):7298. doi: 10.3390/ijms22147298. Int J Mol Sci. 2021. PMID: 34298922 Free PMC article.
-
The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes.Nucleic Acids Res. 2012 Mar;40(6):2683-99. doi: 10.1093/nar/gkr1083. Epub 2011 Nov 28. Nucleic Acids Res. 2012. PMID: 22123745 Free PMC article.
-
RNA interference-mediated silencing of eukaryotic translation initiation factor 3, subunit B (EIF3B) gene expression inhibits proliferation of colon cancer cells.World J Surg Oncol. 2012 Jun 26;10:119. doi: 10.1186/1477-7819-10-119. World J Surg Oncol. 2012. PMID: 22734884 Free PMC article.
-
Heterogeneity and specialized functions of translation machinery: from genes to organisms.Nat Rev Genet. 2018 Jul;19(7):431-452. doi: 10.1038/s41576-018-0008-z. Nat Rev Genet. 2018. PMID: 29725087 Free PMC article. Review.
-
Examining weak protein-protein interactions in start codon recognition via NMR spectroscopy.FEBS J. 2014 Apr;281(8):1965-73. doi: 10.1111/febs.12667. Epub 2014 Jan 2. FEBS J. 2014. PMID: 24393460 Free PMC article. Review.
References
-
- Hinnebusch AG, Dever TE, Asano KA. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. In: Sonenberg N, Mathews M, Hershey JWB, editors. Translational Control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 225–268.
-
- Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Sonenberg N, Mathews M, Hershey JWB, editors. Translational Control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128.
-
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

