Insights into the domain and repeat architecture of target of rapamycin
- PMID: 20060908
- PMCID: PMC2856717
- DOI: 10.1016/j.jsb.2010.01.002
Insights into the domain and repeat architecture of target of rapamycin
Abstract
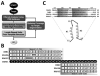
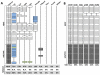
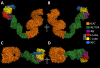
A simple and efficient protein sequence analysis strategy was developed to predict the number and location of structural repeats in the TOR protein. This strategy uses multiple HHpred alignments against proteins of known 3D structure to enable protein repeats referenced from the 3D structure to be traced back to the query protein sequence by using user-directed repeat assignments. The HHpred strategy performed with high sensitivity by predicting 100% of the repeat units within a test set of HEAT- and TPR-repeat-containing proteins of known three-dimensional structure. The HHpred strategy predicts that TOR contains 32 tandem HEAT repeats extending from the N-terminus to the FAT domain, which is itself comprised of 16 tandem TPR repeats. These findings were used to assemble a 3D atomic model for the TOR protein.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Activating mutations in TOR are in similar structures as oncogenic mutations in PI3KCalpha.ACS Chem Biol. 2009 Dec 18;4(12):999-1015. doi: 10.1021/cb900193e. ACS Chem Biol. 2009. PMID: 19902965 Free PMC article.
-
Highly sensitive detection of individual HEAT and ARM repeats with HHpred and COACH.PLoS One. 2009 Sep 24;4(9):e7148. doi: 10.1371/journal.pone.0007148. PLoS One. 2009. PMID: 19777061 Free PMC article.
-
[Bioinformatic search for plant homologs of Ste20-like serine/threonine protein kinases].Tsitol Genet. 2009 Nov-Dec;43(6):68-77. Tsitol Genet. 2009. PMID: 20458979 Russian.
-
TPR proteins: the versatile helix.Trends Biochem Sci. 2003 Dec;28(12):655-62. doi: 10.1016/j.tibs.2003.10.007. Trends Biochem Sci. 2003. PMID: 14659697 Review.
-
The tetratricopeptide repeat: a structural motif mediating protein-protein interactions.Bioessays. 1999 Nov;21(11):932-9. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. Bioessays. 1999. PMID: 10517866 Review.
Cited by
-
Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes.Mol Cell Biol. 2011 Feb;31(4):818-31. doi: 10.1128/MCB.00687-10. Epub 2010 Dec 13. Mol Cell Biol. 2011. PMID: 21149579 Free PMC article.
-
Functional and genomic analyses of alpha-solenoid proteins.PLoS One. 2013 Nov 21;8(11):e79894. doi: 10.1371/journal.pone.0079894. eCollection 2013. PLoS One. 2013. PMID: 24278209 Free PMC article.
-
Plant TOR signaling components.Plant Signal Behav. 2011 Nov;6(11):1700-5. doi: 10.4161/psb.6.11.17662. Epub 2011 Nov 1. Plant Signal Behav. 2011. PMID: 22057328 Free PMC article. Review.
-
Activating mutations of TOR (target of rapamycin).Genes Cells. 2011 Feb;16(2):141-51. doi: 10.1111/j.1365-2443.2010.01482.x. Epub 2011 Jan 7. Genes Cells. 2011. PMID: 21210909 Free PMC article. Review.
-
Emerging Role of mTOR Signaling-Related miRNAs in Cardiovascular Diseases.Oxid Med Cell Longev. 2018 Aug 23;2018:6141902. doi: 10.1155/2018/6141902. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 30305865 Free PMC article. Review.
References
-
- Choi J, et al. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273(5272):239–42. - PubMed
-
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–9. - PubMed
-
- Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73(3):585–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

