Insights into the domain and repeat architecture of target of rapamycin
- PMID: 20060908
- PMCID: PMC2856717
- DOI: 10.1016/j.jsb.2010.01.002
Insights into the domain and repeat architecture of target of rapamycin
Abstract
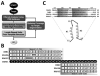
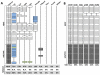
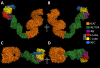
A simple and efficient protein sequence analysis strategy was developed to predict the number and location of structural repeats in the TOR protein. This strategy uses multiple HHpred alignments against proteins of known 3D structure to enable protein repeats referenced from the 3D structure to be traced back to the query protein sequence by using user-directed repeat assignments. The HHpred strategy performed with high sensitivity by predicting 100% of the repeat units within a test set of HEAT- and TPR-repeat-containing proteins of known three-dimensional structure. The HHpred strategy predicts that TOR contains 32 tandem HEAT repeats extending from the N-terminus to the FAT domain, which is itself comprised of 16 tandem TPR repeats. These findings were used to assemble a 3D atomic model for the TOR protein.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



References
-
- Choi J, et al. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273(5272):239–42. - PubMed
-
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–9. - PubMed
-
- Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73(3):585–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

