Genome organization and characterization of the virulent lactococcal phage 1358 and its similarities to Listeria phages
- PMID: 20061452
- PMCID: PMC2832367
- DOI: 10.1128/AEM.02173-09
Genome organization and characterization of the virulent lactococcal phage 1358 and its similarities to Listeria phages
Abstract
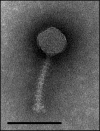
Virulent phage 1358 is the reference member of a rare group of phages infecting Lactococcus lactis. Electron microscopy revealed a typical icosahedral capsid connected to one of the smallest noncontractile tails found in a lactococcal phage of the Siphoviridae family. Microbiological characterization identified a burst size of 72 virions released per infected host cell and a latent period of 90 min. The host range of phage 1358 was limited to 3 out of the 60 lactococcal strains tested. Moreover, this phage was insensitive to four Abi systems (AbiK, AbiQ, AbiT, and AbiV). The genome of phage 1358 consisted of a linear, double-stranded, 36,892-bp DNA molecule containing 43 open reading frames (ORFs). At least 14 ORFs coded for structural proteins, as identified by SDS-PAGE coupled to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. The genomic organization was similar to those of other siphophages. All genes were on the same coding strand and in the same orientation. This lactococcal phage was unique, however, in its 51.4% GC content, much higher than those of other phages infecting this low-GC Gram-positive host. A bias for GC-rich codons was also observed. Comparative analyses showed that several phage 1358 structural proteins shared similarity with two Listeria monocytogenes phages, P35 and P40. The possible origin and evolution of lactococcal phage 1358 is discussed.
Figures


Similar articles
-
Cryo-electron microscopy structure of lactococcal siphophage 1358 virion.J Virol. 2014 Aug;88(16):8900-10. doi: 10.1128/JVI.01040-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872584 Free PMC article.
-
Characterization of Lactococcus lactis phage 949 and comparison with other lactococcal phages.Appl Environ Microbiol. 2010 Oct;76(20):6843-52. doi: 10.1128/AEM.00796-10. Epub 2010 Aug 27. Appl Environ Microbiol. 2010. PMID: 20802084 Free PMC article.
-
A virulent phage infecting Lactococcus garvieae, with homology to Lactococcus lactis phages.Appl Environ Microbiol. 2015 Dec;81(24):8358-65. doi: 10.1128/AEM.02603-15. Epub 2015 Sep 25. Appl Environ Microbiol. 2015. PMID: 26407890 Free PMC article.
-
Structures and host-adhesion mechanisms of lactococcal siphophages.Front Microbiol. 2014 Jan 16;5:3. doi: 10.3389/fmicb.2014.00003. eCollection 2014. Front Microbiol. 2014. PMID: 24474948 Free PMC article. Review.
-
Origin, Evolution and Diversity of φ29-like Phages-Review and Bioinformatic Analysis.Int J Mol Sci. 2024 Oct 9;25(19):10838. doi: 10.3390/ijms251910838. Int J Mol Sci. 2024. PMID: 39409167 Free PMC article. Review.
Cited by
-
Bacteriophages and dairy fermentations.Bacteriophage. 2012 Jul 1;2(3):149-158. doi: 10.4161/bact.21868. Bacteriophage. 2012. PMID: 23275866 Free PMC article.
-
Cryo-electron microscopy structure of lactococcal siphophage 1358 virion.J Virol. 2014 Aug;88(16):8900-10. doi: 10.1128/JVI.01040-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872584 Free PMC article.
-
Characterization of the First Virulent Phage Infecting Oenococcus oeni, the Queen of the Cellars.Front Microbiol. 2021 Jan 13;11:596541. doi: 10.3389/fmicb.2020.596541. eCollection 2020. Front Microbiol. 2021. PMID: 33519734 Free PMC article.
-
Global Survey and Genome Exploration of Bacteriophages Infecting the Lactic Acid Bacterium Streptococcus thermophilus.Front Microbiol. 2017 Sep 12;8:1754. doi: 10.3389/fmicb.2017.01754. eCollection 2017. Front Microbiol. 2017. PMID: 28955321 Free PMC article.
-
Codon Optimization is Required to Express Fluorogenic Reporter Proteins in Lactococcus lactis.Mol Biotechnol. 2024 Sep 23. doi: 10.1007/s12033-024-01285-5. Online ahead of print. Mol Biotechnol. 2024. PMID: 39312082
References
-
- Ackermann, H.-W., G. Brochu, and H. P. E. Konjin. 1994. Classification of Acinetobacter phages. Arch. Virol. 135:345-354. - PubMed
-
- Ackermann, H.-W., and A. M. Kropinski. 2007. Curated list of prokaryote viruses with fully sequenced genomes. Res. Microbiol. 158:555-566. - PubMed
-
- Allison, G. E., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

