Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium
- PMID: 20061468
- PMCID: PMC2832528
- DOI: 10.1128/JB.01518-09
Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium
Abstract
To gain insights into the origin and genome evolution of the plague bacterium Yersinia pestis, we have sequenced the deep-rooted strain Angola, a virulent Pestoides isolate. Its ancient nature makes this atypical isolate of particular importance in understanding the evolution of plague pathogenicity. Its chromosome features a unique genetic make-up intermediate between modern Y. pestis isolates and its evolutionary ancestor, Y. pseudotuberculosis. Our genotypic and phenotypic analyses led us to conclude that Angola belongs to one of the most ancient Y. pestis lineages thus far sequenced. The mobilome carries the first reported chimeric plasmid combining the two species-specific virulence plasmids. Genomic findings were validated in virulence assays demonstrating that its pathogenic potential is distinct from modern Y. pestis isolates. Human infection with this particular isolate would not be diagnosed by the standard clinical tests, as Angola lacks the plasmid-borne capsule, and a possible emergence of this genotype raises major public health concerns. To assess the genomic plasticity in Y. pestis, we investigated the global gene reservoir and estimated the pangenome at 4,844 unique protein-coding genes. As shown by the genomic analysis of this evolutionary key isolate, we found that the genomic plasticity within Y. pestis clearly was not as limited as previously thought, which is strengthened by the detection of the largest number of isolate-specific single-nucleotide polymorphisms (SNPs) currently reported in the species. This study identified numerous novel genetic signatures, some of which seem to be intimately associated with plague virulence. These markers are valuable in the development of a robust typing system critical for forensic, diagnostic, and epidemiological studies.
Figures
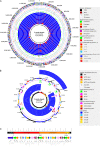

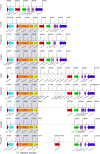
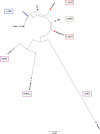
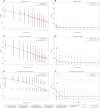
Similar articles
-
Pestoides F, an atypical Yersinia pestis strain from the former Soviet Union.Adv Exp Med Biol. 2007;603:17-22. doi: 10.1007/978-0-387-72124-8_2. Adv Exp Med Biol. 2007. PMID: 17966401
-
Microevolution and history of the plague bacillus, Yersinia pestis.Proc Natl Acad Sci U S A. 2004 Dec 21;101(51):17837-42. doi: 10.1073/pnas.0408026101. Epub 2004 Dec 14. Proc Natl Acad Sci U S A. 2004. PMID: 15598742 Free PMC article.
-
Novel plasmids and resistance phenotypes in Yersinia pestis: unique plasmid inventory of strain Java 9 mediates high levels of arsenic resistance.PLoS One. 2012;7(3):e32911. doi: 10.1371/journal.pone.0032911. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479347 Free PMC article.
-
Plague in the genomic area.Clin Microbiol Infect. 2012 Mar;18(3):224-30. doi: 10.1111/j.1469-0691.2012.03774.x. Clin Microbiol Infect. 2012. PMID: 22369155 Review.
-
Review of genotyping methods for Yersinia pestis in Madagascar.PLoS Negl Trop Dis. 2024 Jun 27;18(6):e0012252. doi: 10.1371/journal.pntd.0012252. eCollection 2024 Jun. PLoS Negl Trop Dis. 2024. PMID: 38935608 Free PMC article. Review.
Cited by
-
Averting Behavior Framework for Perceived Risk of Yersinia enterocolitica Infections.J Pathog. 2012;2012:725373. doi: 10.1155/2012/725373. Epub 2012 Mar 7. J Pathog. 2012. PMID: 22619726 Free PMC article.
-
Rapid Detection of Genetic Engineering, Structural Variation, and Antimicrobial Resistance Markers in Bacterial Biothreat Pathogens by Nanopore Sequencing.Sci Rep. 2019 Sep 18;9(1):13501. doi: 10.1038/s41598-019-49700-1. Sci Rep. 2019. PMID: 31534162 Free PMC article.
-
Genetic and Virulence Profiles of Enteroaggregative Escherichia coli (EAEC) Isolated From Deployed Military Personnel (DMP) With Travelers' Diarrhea.Front Cell Infect Microbiol. 2020 May 20;10:200. doi: 10.3389/fcimb.2020.00200. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32509590 Free PMC article.
-
Pathogenomes of Atypical Non-shigatoxigenic Escherichia coli NSF/SF O157:H7/NM: Comprehensive Phylogenomic Analysis Using Closed Genomes.Front Microbiol. 2020 Apr 15;11:619. doi: 10.3389/fmicb.2020.00619. eCollection 2020. Front Microbiol. 2020. PMID: 32351476 Free PMC article.
-
Short and long-term genome stability analysis of prokaryotic genomes.BMC Genomics. 2013 May 8;14:309. doi: 10.1186/1471-2164-14-309. BMC Genomics. 2013. PMID: 23651581 Free PMC article.
References
-
- Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 101:17837-17842. - PMC - PubMed
-
- Adamovicz, J. L., and P. L. Worsham. 2006. Plague, p. 107-135. In J. R. Swearengen (ed.), Biodefense: research methodology and animal models. CRC Press Taylor & Francis Group, Boca Raton, FL.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

