PepD participates in the mycobacterial stress response mediated through MprAB and SigE
- PMID: 20061478
- PMCID: PMC2832534
- DOI: 10.1128/JB.01167-09
PepD participates in the mycobacterial stress response mediated through MprAB and SigE
Abstract
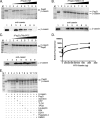
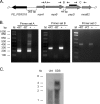
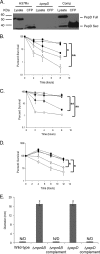
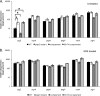
Currently, one-third of the world's population is believed to be latently infected with Mycobacterium tuberculosis. The mechanisms by which M. tuberculosis establishes latent infection remain largely undefined. mprAB encodes a two-component signal transduction system required by M. tuberculosis for aspects of persistent infection. MprAB regulates a large and diverse group of genetic determinants in response to membrane stress, including the extracytoplasmic function (ECF) sigma factor sigE and the HtrA-like serine protease pepD. Recent studies have demonstrated that PepD functions as both a protease and chaperone in vitro. In addition, inactivation of pepD alters the virulence of M. tuberculosis in a mouse model system of infection. Here, we demonstrate that PepD plays an important role in the stress response network of Mycobacterium mediated through MprAB and SigE. In particular, we demonstrate that the protease activity of PepD requires the PDZ domain, in addition to the catalytic serine at position 317. pepD expression initiates from at least three promoters in M. tuberculosis, including one that is regulated by SigE and is located upstream of the mprA coding sequence. Deletion of pepD or mprAB in Mycobacterium smegmatis and M. tuberculosis alters the stress response phenotypes of these strains, including increasing sensitivity to SDS and cell wall antibiotics and upregulating the expression of stress-responsive determinants, including sigE. Taking these data together, we hypothesize that PepD utilizes its PDZ domain to recognize and process misfolded proteins at the cell membrane, leading to activation of the MprAB and SigE signaling pathways and subsequent establishment of a positive feedback loop that facilitates bacterial adaptation.
Figures



References
-
- Alba, B. M., H. J. Zhong, J. C. Pelayo, and C. A. Gross. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma (E) activity. Mol. Microbiol. 40:1323-1333. - PubMed
-
- Anonymous. 2008. Cycloserine. Tuberculosis (Edinb.) 88:100-101. - PubMed
-
- Anonymous. 2008. Global tuberculosis control. Surveillance, planning, financing. World Health Organization, Geneva, Switzerland.
-
- Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

