Global change of gene expression and cell physiology in YidC-depleted Escherichia coli
- PMID: 20061485
- PMCID: PMC2849450
- DOI: 10.1128/JB.00484-09
Global change of gene expression and cell physiology in YidC-depleted Escherichia coli
Abstract
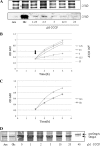
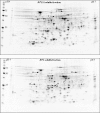
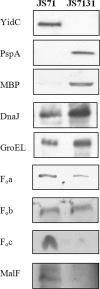
YidC depletion affects membrane protein insertion and leads to a defect in the growth of the Escherichia coli cell. We analyzed global changes in gene expression upon YidC depletion to determine the importance of YidC for cellular functions using a gene chip method to compare the transcriptomes of JS71 (control) and JS7131 (yidC depletion strain). Of the more than 4,300 genes identified, 163 were upregulated and 99 were downregulated upon YidC depletion, including genes which are responsible for DNA/RNA repair; energy metabolism; various transporters, proteases and chaperones; stress response; and translation and transcription functions. Real-time PCR was performed on selected genes to confirm the results. Specifically, we found upregulation of the genes encoding the energy transduction proteins F(1)F(o) ATP synthase and cytochrome bo(3) oxidase due to perturbation in assembly when YidC was depleted. We also determined that the high-level induction of the PspA stress protein under YidC depletion conditions is roughly 10-fold higher than the activation due to the addition of protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP), which dissipates the proton motive force. In addition, the gene chip data reveal the Cpx stress pathway is activated upon YidC depletion. The data show the broad physiological contribution of YidC to the bacterial cell and the considerable ramification to the cell when it is depleted.
Figures

Similar articles
-
Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs.J Bacteriol. 2008 Apr;190(7):2458-69. doi: 10.1128/JB.01366-07. Epub 2008 Jan 4. J Bacteriol. 2008. PMID: 18178746 Free PMC article.
-
YidC is involved in the biogenesis of anaerobic respiratory complexes in the inner membrane of Escherichia coli.J Biol Chem. 2008 Oct 3;283(40):26921-7. doi: 10.1074/jbc.M804490200. Epub 2008 Jul 17. J Biol Chem. 2008. PMID: 18635537
-
Characterization of the consequences of YidC depletion on the inner membrane proteome of E. coli using 2D blue native/SDS-PAGE.J Mol Biol. 2011 Jun 3;409(2):124-35. doi: 10.1016/j.jmb.2011.03.068. Epub 2011 Apr 8. J Mol Biol. 2011. PMID: 21497606
-
Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes.J Biol Chem. 2008 Nov 14;283(46):31269-73. doi: 10.1074/jbc.R800029200. Epub 2008 Jul 25. J Biol Chem. 2008. PMID: 18658156 Review.
-
YidC-mediated membrane insertion.FEMS Microbiol Lett. 2018 Jun 1;365(12). doi: 10.1093/femsle/fny106. FEMS Microbiol Lett. 2018. PMID: 29800285 Review.
Cited by
-
Membrane proteomic analysis reveals overlapping and independent functions of Streptococcus mutans Ffh, YidC1, and YidC2.Mol Oral Microbiol. 2019 Aug;34(4):131-152. doi: 10.1111/omi.12261. Epub 2019 Jun 7. Mol Oral Microbiol. 2019. PMID: 31034136 Free PMC article.
-
Each protomer of a dimeric YidC functions as a single membrane insertase.Sci Rep. 2018 Jan 12;8(1):589. doi: 10.1038/s41598-017-18830-9. Sci Rep. 2018. PMID: 29330366 Free PMC article.
-
Phage shock proteins B and C prevent lethal cytoplasmic membrane permeability in Yersinia enterocolitica.Mol Microbiol. 2012 Aug;85(3):445-60. doi: 10.1111/j.1365-2958.2012.08120.x. Epub 2012 Jun 12. Mol Microbiol. 2012. PMID: 22646656 Free PMC article.
-
Dynamic disulfide scanning of the membrane-inserting Pf3 coat protein reveals multiple YidC substrate contacts.J Biol Chem. 2012 Feb 3;287(6):3769-76. doi: 10.1074/jbc.M111.307223. Epub 2011 Dec 16. J Biol Chem. 2012. PMID: 22179606 Free PMC article.
-
Biogenesis of bacterial inner-membrane proteins.Cell Mol Life Sci. 2010 Jul;67(14):2343-62. doi: 10.1007/s00018-010-0303-0. Epub 2010 Mar 5. Cell Mol Life Sci. 2010. PMID: 20204450 Free PMC article. Review.
References
-
- Celebi, N., L. Yi, S. J. Facey, A. Kuhn, and R. E. Dalbey. 2006. Membrane biogenesis of subunit II of cytochrome bo oxidase: contrasting requirements for insertion of N-terminal and C-terminal domains. J. Mol. Biol. 357:1428-1436. - PubMed
-
- Chen, M., K. Xie, J. Yuan, L. Yi, S. J. Facey, N. Pradel, L. F. Wu, A. Kuhn, and R. E. Dalbey. 2005. Involvement of SecDF and YidC in the membrane insertion of M13 procoat mutants. Biochemistry 44:10741-10749. - PubMed
-
- Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

