Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time
- PMID: 20061554
- PMCID: PMC2828697
- DOI: 10.1105/tpc.109.065490
Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time
Abstract
CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) possesses E3 ligase activity and promotes degradation of key factors involved in the light regulation of plant development. The finding that CULLIN4 (CUL4)-Damaged DNA Binding Protein1 (DDB1) interacts with DDB1 binding WD40 (DWD) proteins to act as E3 ligases implied that CUL4-DDB1 may associate with COP1-SUPPRESSOR OF PHYA (SPA) protein complexes, since COP1 and SPAs are DWD proteins. Here, we demonstrate that CUL4-DDB1 physically associates with COP1-SPA complexes in vitro and in vivo, likely via direct interaction of DDB1 with COP1 and SPAs. The interactions between DDB1 and COP1, SPA1, and SPA3 were disrupted by mutations in the WDXR motifs of MBP-COP1, His-SPA1, and His-SPA3. CUL4 cosuppression mutants enhanced weak cop1 photomorphogenesis and flowered early under short days. Early flowering of short day-grown cul4 mutants correlated with increased FLOWERING LOCUS T transcript levels, whereas CONSTANS transcript levels were not altered. De-etiolated1 and COP1 can bind DDB1 and may work with CUL4-DDB1 in distinct complexes, but they mediate photomorphogenesis in concert. Thus, a series of CUL4-DDB1-COP1-SPA E3 ligase complexes may mediate the repression of photomorphogenesis and, possibly, of flowering time.
Figures
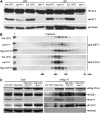
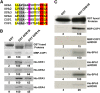


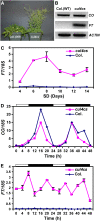

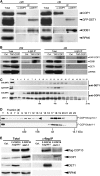

Similar articles
-
Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes.Plant Cell. 2008 Jun;20(6):1437-55. doi: 10.1105/tpc.108.058891. Epub 2008 Jun 13. Plant Cell. 2008. PMID: 18552200 Free PMC article.
-
Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B.Proc Natl Acad Sci U S A. 2013 Oct 8;110(41):16669-74. doi: 10.1073/pnas.1316622110. Epub 2013 Sep 25. Proc Natl Acad Sci U S A. 2013. PMID: 24067658 Free PMC article.
-
Genetic interactions of Arabidopsis thaliana damaged DNA binding protein 1B (DDB1B) with DDB1A, DET1, and COP1.G3 (Bethesda). 2013 Mar;3(3):493-503. doi: 10.1534/g3.112.005249. Epub 2013 Mar 1. G3 (Bethesda). 2013. PMID: 23450167 Free PMC article.
-
The photomorphogenic repressors COP1 and DET1: 20 years later.Trends Plant Sci. 2012 Oct;17(10):584-93. doi: 10.1016/j.tplants.2012.05.004. Epub 2012 Jun 15. Trends Plant Sci. 2012. PMID: 22705257 Review.
-
Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase.Curr Opin Plant Biol. 2018 Oct;45(Pt A):18-25. doi: 10.1016/j.pbi.2018.04.018. Epub 2018 May 15. Curr Opin Plant Biol. 2018. PMID: 29775763 Review.
Cited by
-
The COP1 E3-ligase interacts with FIP200, a key regulator of mammalian autophagy.BMC Biochem. 2013 Jan 6;14:1. doi: 10.1186/1471-2091-14-1. BMC Biochem. 2013. PMID: 23289756 Free PMC article.
-
OsCBE1, a Substrate Receptor of Cullin4-Based E3 Ubiquitin Ligase, Functions as a Regulator of Abiotic Stress Response and Productivity in Rice.Int J Mol Sci. 2021 Mar 2;22(5):2487. doi: 10.3390/ijms22052487. Int J Mol Sci. 2021. PMID: 33801226 Free PMC article.
-
The De-Etiolated 1 Homolog of Arabidopsis Modulates the ABA Signaling Pathway and ABA Biosynthesis in Rice.Plant Physiol. 2016 Jun;171(2):1259-76. doi: 10.1104/pp.16.00059. Epub 2016 May 2. Plant Physiol. 2016. PMID: 27208292 Free PMC article.
-
CRY arrests Cop1 to regulate circadian rhythms in mammals.Cell Div. 2019 Nov 2;14:12. doi: 10.1186/s13008-019-0055-7. eCollection 2019. Cell Div. 2019. PMID: 31700528 Free PMC article.
-
Ethylene-independent promotion of photomorphogenesis in the dark by cytokinin requires COP1 and the CDD complex.J Exp Bot. 2019 Jan 1;70(1):165-178. doi: 10.1093/jxb/ery344. J Exp Bot. 2019. PMID: 30272197 Free PMC article.
References
-
- An H., Roussot C., Suarez-Lopez P., Corbesier L., Vincent C., Pineiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 - PubMed
-
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222 - PubMed
-
- Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., Zheng N. (2006). Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443: 590–593 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

