Riboswitch RNAs: regulation of gene expression by direct monitoring of a physiological signal
- PMID: 20061810
- PMCID: PMC4959611
- DOI: 10.4161/rna.7.1.10757
Riboswitch RNAs: regulation of gene expression by direct monitoring of a physiological signal
Abstract
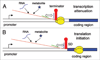
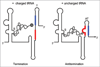
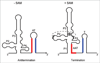
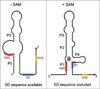
Riboswitches are cis-encoded, cis-acting RNA elements that directly sense a physiological signal. Signal response results in a change in RNA structure that impacts gene expression. Elements of this type play an important role in bacteria, where they regulate a variety of fundamental cellular pathways. Riboswitch-mediated gene regulation most commonly occurs by effects on transcription attenuation, to control whether a full-length transcript is synthesized, or on translation initiation, in which case the transcript is constitutively synthesized but binding of the translation initiation complex is modulated. An overview of the role of riboswitch RNAs in bacterial gene expression will be provided, and a few examples are described in more detail to illustrate the types of mechanisms that have been uncovered.
Figures
References
-
- Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol. 2006;41:329–338. - PubMed
-
- Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. - PubMed
-
- Storz G, Opdyke JA, Wassarman KM. Regulating bacterial transcription with small RNAs. Cold Spring Harbor Symp Quant Biol. 2006;71:269–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
