Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk
- PMID: 20062064
- PMCID: PMC5321238
- DOI: 10.1038/ng.510
Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk
Abstract
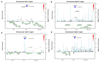
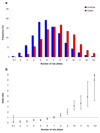
To identify new risk variants for chronic lymphocytic leukemia (CLL), we conducted a genome-wide association study of 299,983 tagging SNPs, with validation in four additional series totaling 2,503 cases and 5,789 controls. We identified four new risk loci for CLL at 2q37.3 (rs757978, FARP2; odds ratio (OR) = 1.39; P = 2.11 x 10(-9)), 8q24.21 (rs2456449; OR = 1.26; P = 7.84 x 10(-10)), 15q21.3 (rs7169431; OR = 1.36; P = 4.74 x 10(-7)) and 16q24.1 (rs305061; OR = 1.22; P = 3.60 x 10(-7)). We also found evidence for risk loci at 15q25.2 (rs783540, CPEB1; OR = 1.18; P = 3.67 x 10(-6)) and 18q21.1 (rs1036935; OR = 1.22; P = 2.28 x 10(-6)). These data provide further evidence for genetic susceptibility to this B-cell hematological malignancy.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104:1850–4. - PubMed
-
- Di Bernardo MC, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–10. - PubMed
-
- Gunnarsson R, et al. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia--a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer. 2008;47:697–711. - PubMed
-
- Ferreira BI, et al. Comparative genome profiling across subtypes of low-grade B-cell lymphoma identifies type-specific and common aberrations that target genes with a role in B-cell neoplasia. Haematologica. 2008;93:670–9. - PubMed
-
- Grubor V, et al. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA) Blood. 2009;113:1294–303. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

