The first bite--profiling the predatosome in the bacterial pathogen Bdellovibrio
- PMID: 20062540
- PMCID: PMC2797640
- DOI: 10.1371/journal.pone.0008599
The first bite--profiling the predatosome in the bacterial pathogen Bdellovibrio
Abstract
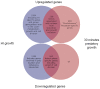
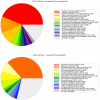
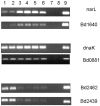
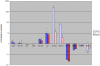
Bdellovibrio bacteriovorus is a Gram-negative bacterium that is a pathogen of other Gram-negative bacteria, including many bacteria which are pathogens of humans, animals and plants. As such Bdellovibrio has potential as a biocontrol agent, or living antibiotic. B. bacteriovorus HD100 has a large genome and it is not yet known which of it encodes the molecular machinery and genetic control of predatory processes. We have tried to fill this knowledge-gap using mixtures of predator and prey mRNAs to monitor changes in Bdellovibrio gene expression at a timepoint of early-stage prey infection and prey killing in comparison to control cultures of predator and prey alone and also in comparison to Bdellovibrio growing axenically (in a prey-or host independent "HI" manner) on artificial media containing peptone and tryptone. From this we have highlighted genes of the early predatosome with predicted roles in prey killing and digestion and have gained insights into possible regulatory mechanisms as Bdellovibrio enter and establish within the prey bdelloplast. Approximately seven percent of all Bdellovibrio genes were significantly up-regulated at 30 minutes of infection--but not in HI growth--implicating the role of these genes in prey digestion. Five percent were down-regulated significantly, implicating their role in free-swimming, attack-phase physiology. This study gives the first post-genomic insight into the predatory process and reveals some of the important genes that Bdellovibrio expresses inside the prey bacterium during the initial attack.
Conflict of interest statement
Figures

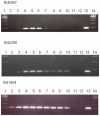


Similar articles
-
Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria.BMC Genomics. 2012 Nov 27;13:670. doi: 10.1186/1471-2164-13-670. BMC Genomics. 2012. PMID: 23181807 Free PMC article.
-
Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus.PLoS Pathog. 2012 Feb;8(2):e1002493. doi: 10.1371/journal.ppat.1002493. Epub 2012 Feb 2. PLoS Pathog. 2012. PMID: 22319440 Free PMC article.
-
Downregulation of the motA gene delays the escape of the obligate predator Bdellovibrio bacteriovorus 109J from bdelloplasts of bacterial prey cells.Microbiology (Reading). 2004 Mar;150(Pt 3):649-656. doi: 10.1099/mic.0.26761-0. Microbiology (Reading). 2004. PMID: 14993314
-
A predatory patchwork: membrane and surface structures of Bdellovibrio bacteriovorus.Adv Microb Physiol. 2009;54:313-61. doi: 10.1016/S0065-2911(08)00005-2. Adv Microb Physiol. 2009. PMID: 18929071 Review.
-
Predatory lifestyle of Bdellovibrio bacteriovorus.Annu Rev Microbiol. 2009;63:523-39. doi: 10.1146/annurev.micro.091208.073346. Annu Rev Microbiol. 2009. PMID: 19575566 Review.
Cited by
-
A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus.PLoS One. 2013 Apr 16;8(4):e61850. doi: 10.1371/journal.pone.0061850. Print 2013. PLoS One. 2013. PMID: 23613952 Free PMC article.
-
A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus.Nat Commun. 2020 Sep 23;11(1):4817. doi: 10.1038/s41467-020-18139-8. Nat Commun. 2020. PMID: 32968056 Free PMC article.
-
Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness.PLoS Pathog. 2012 Feb;8(2):e1002524. doi: 10.1371/journal.ppat.1002524. Epub 2012 Feb 9. PLoS Pathog. 2012. PMID: 22346754 Free PMC article.
-
Using AlphaFold-Multimer to study novel protein-protein interactions of predation essential hypothetical proteins in Bdellovibrio.Front Bioinform. 2025 Apr 14;5:1566486. doi: 10.3389/fbinf.2025.1566486. eCollection 2025. Front Bioinform. 2025. PMID: 40297267 Free PMC article.
-
DivIVA Controls Progeny Morphology and Diverse ParA Proteins Regulate Cell Division or Gliding Motility in Bdellovibrio bacteriovorus.Front Microbiol. 2020 Apr 21;11:542. doi: 10.3389/fmicb.2020.00542. eCollection 2020. Front Microbiol. 2020. PMID: 32373080 Free PMC article.
References
-
- Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. - PubMed
-
- Lambert C, Sockett RE. Laboratory maintenance of Bdellovibrio. Curr Protoc Microbiol Chapter. 2008;7:Unit 7B 2. - PubMed
-
- Saeed AI, Sharov V, White J, Li J, Liang W, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

