GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex
- PMID: 20063437
- PMCID: PMC2805832
- DOI: 10.1002/prot.22507
GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex
Abstract
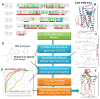
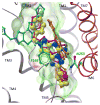
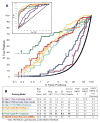
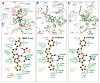
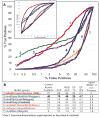
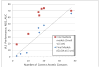
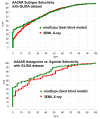
Proteins of the G-protein coupled receptor (GPCR) family present numerous attractive targets for rational drug design, but also a formidable challenge for identification and conformational modeling of their 3D structure. A recently performed assessment of blind predictions of adenosine A2a receptor (AA2AR) structure in complex with ZM241385 (ZMA) antagonist provided a first example of unbiased evaluation of the current modeling algorithms on a GPCR target with approximately 30% sequence identity to the closest structural template. Several of the 29 groups participating in this assessment exercise (Michino et al., doi: 10.1038/nrd2877) successfully predicted the overall position of the ligand ZMA in the AA2AR ligand binding pocket, however models from only three groups captured more than 40% the ligand-receptor contacts. Here we describe two of these top performing approaches, in which all-atom models of the AA2AR were generated by homology modeling followed by ligand guided backbone ensemble receptor optimization (LiBERO). The resulting AA2AR-ZMA models, along with the best models from other groups are assessed here for their vitual ligand screening (VLS) performance on a large set of GPCR ligands. We show that ligand guided optimization was critical for improvement of both ligand-receptor contacts and VLS performance as compared to the initial raw homology models. The best blindly predicted models performed on par with the crystal structure of AA2AR in selecting known antagonists from decoys, as well as from antagonists for other adenosine subtypes and AA2AR agonists. These results suggest that despite certain inaccuracies, the optimized homology models can be useful in the drug discovery process.
(c) 2009 Wiley-Liss, Inc.
Figures

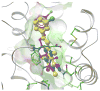
References
-
- Reynolds K, Abagyan R, Katritch V. 3d structure and modeling of GPCRs: implications for drug discovery. In: Gilchrist A, editor. Shifting Paradigms in G Protein Coupled Receptors. Hoboken, NJ: Wiley & Sons, Inc; 2009. in press.
-
- Tyndall JD, Sandilya R. GPCR agonists and antagonists in the clinic. Med Chem. 2005;1(4):405–421. - PubMed
-
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7(4):339–357. - PubMed
-
- Jacoby E, Bouhelal R, Gerspacher M, Seuwen K. The 7 TM G-protein-coupled receptor target family. ChemMedChem. 2006;1(8):761–782. - PubMed
-
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

