Effects of interdomain tether length and flexibility on the kinetics of intramolecular electron transfer in human sulfite oxidase
- PMID: 20063894
- PMCID: PMC2819551
- DOI: 10.1021/bi9020296
Effects of interdomain tether length and flexibility on the kinetics of intramolecular electron transfer in human sulfite oxidase
Abstract
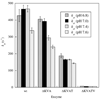
Sulfite oxidase (SO) is a vitally important molybdenum enzyme that catalyzes the oxidation of toxic sulfite to sulfate. The proposed catalytic mechanism of vertebrate SO involves two intramolecular one-electron transfer (IET) steps from the molybdenum cofactor to the iron of the integral b-type heme and two intermolecular one-electron steps to exogenous cytochrome c. In the crystal structure of chicken SO [Kisker, C., et al. (1997) Cell 91, 973-983], which is highly homologous to human SO (HSO), the heme iron and molybdenum centers are separated by 32 A and the domains containing these centers are linked by a flexible polypeptide tether. Conformational changes that bring these two centers into greater proximity have been proposed [Feng, C., et al. (2003) Biochemistry 42, 5816-5821] to explain the relatively rapid IET kinetics, which are much faster than those theoretically predicted from the crystal structure. To explore the proposed role(s) of the tether in facilitating this conformational change, we altered both its length and flexibility in HSO by site-specific mutagenesis, and the reactivities of the resulting variants have been studied using laser flash photolysis and steady-state kinetics assays. Increasing the flexibility of the tether by mutating several conserved proline residues to alanines did not produce a discernible systematic trend in the kinetic parameters, although mutation of one residue (P105) to alanine produced a 3-fold decrease in the IET rate constant. Deletions of nonconserved amino acids in the 14-residue tether, thereby shortening its length, resulted in more drastically reduced IET rate constants. Thus, the deletion of five amino acid residues decreased IET by 70-fold, so that it was rate-limiting in the overall reaction. The steady-state kinetic parameters were also significantly affected by these mutations, with the P111A mutation causing a 5-fold increase in the sulfite K(m) value, perhaps reflecting a decrease in the ability to bind sulfite. The electron paramagnetic resonance spectra of these proline to alanine and deletion mutants are identical to those of wild-type HSO, indicating no significant change in the Mo active site geometry.
Figures
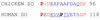


Similar articles
-
Elucidating the catalytic mechanism of sulfite oxidizing enzymes using structural, spectroscopic, and kinetic analyses.Biochemistry. 2010 Aug 31;49(34):7242-54. doi: 10.1021/bi1008485. Biochemistry. 2010. PMID: 20666399 Free PMC article. Review.
-
Effects of large-scale amino acid substitution in the polypeptide tether connecting the heme and molybdenum domains on catalysis in human sulfite oxidase.Metallomics. 2010 Nov;2(11):766-70. doi: 10.1039/c0mt00021c. Epub 2010 Sep 20. Metallomics. 2010. PMID: 21072368
-
Effects of mutating aromatic surface residues of the heme domain of human sulfite oxidase on its heme midpoint potential, intramolecular electron transfer, and steady-state kinetics.Dalton Trans. 2013 Mar 7;42(9):3043-9. doi: 10.1039/c2dt31508d. Epub 2012 Sep 14. Dalton Trans. 2013. PMID: 22975842
-
Mechanistic complexities of sulfite oxidase: An enzyme with multiple domains, subunits, and cofactors.J Inorg Biochem. 2023 Oct;247:112312. doi: 10.1016/j.jinorgbio.2023.112312. Epub 2023 Jul 4. J Inorg Biochem. 2023. PMID: 37441922
-
Sulfite oxidizing enzymes.Biochim Biophys Acta. 2007 May;1774(5):527-39. doi: 10.1016/j.bbapap.2007.03.006. Epub 2007 Mar 20. Biochim Biophys Acta. 2007. PMID: 17459792 Free PMC article. Review.
Cited by
-
Hydrogen Sulfide Oxidation by Sulfide Quinone Oxidoreductase.Chembiochem. 2021 Mar 16;22(6):949-960. doi: 10.1002/cbic.202000661. Epub 2020 Nov 17. Chembiochem. 2021. PMID: 33080111 Free PMC article. Review.
-
Nitrite reduction by molybdoenzymes: a new class of nitric oxide-forming nitrite reductases.J Biol Inorg Chem. 2015 Mar;20(2):403-33. doi: 10.1007/s00775-014-1234-2. Epub 2015 Jan 15. J Biol Inorg Chem. 2015. PMID: 25589250 Review.
-
Another look at the interaction between mitochondrial cytochrome c and flavocytochrome b (2).Eur Biophys J. 2011 Dec;40(12):1283-99. doi: 10.1007/s00249-011-0697-0. Epub 2011 Apr 19. Eur Biophys J. 2011. PMID: 21503671 Review.
-
Kinetic results for mutations of conserved residues H304 and R309 of human sulfite oxidase point to mechanistic complexities.Metallomics. 2014 Sep;6(9):1664-70. doi: 10.1039/c4mt00099d. Metallomics. 2014. PMID: 24968320 Free PMC article.
-
Elucidating the catalytic mechanism of sulfite oxidizing enzymes using structural, spectroscopic, and kinetic analyses.Biochemistry. 2010 Aug 31;49(34):7242-54. doi: 10.1021/bi1008485. Biochemistry. 2010. PMID: 20666399 Free PMC article. Review.
References
-
- Hille R. The Mononuclear Molybdenum Enzymes. Chem. Rev. 1996;96:2757–2816. - PubMed
-
- Rajagopalan KV, Johnson JL. Sulfite Oxidase. In: Creighton TE, editor. Wiley Encyclopedia of Molecular Medicine. New York: Wiley; 2002. pp. 3048–3051.
-
- Kisker C. Sulfite Oxidase. In: Messerschmidt A, Huber R, Poulos T, Wieghardt K, editors. Handbook of Metalloproteins. New York: John Wiley and Sons, Ltd; 2001. pp. 1121–1135.
-
- Schindelin H, Kisker C, Rajagopalan KV. Molybdopterin from Molybdenum and Tungsten Enzymes. Adv. Protein Chem. 2001;58:47–94. - PubMed
-
- Cohen HJ, Betcher-Lange S, Kessler DL, Rajagopalan KV. Hepatic Sulfite Oxidase. Congruency in Mitochondria of Prosthetic Groups and Activity. J. Biol. Chem. 1972;247:7759–7766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

