Integrating phenotype ontologies across multiple species
- PMID: 20064205
- PMCID: PMC2847714
- DOI: 10.1186/gb-2010-11-1-r2
Integrating phenotype ontologies across multiple species
Abstract
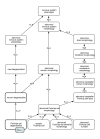
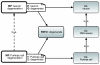
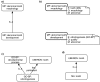
Phenotype ontologies are typically constructed to serve the needs of a particular community, such as annotation of genotype-phenotype associations in mouse or human. Here we demonstrate how these ontologies can be improved through assignment of logical definitions using a core ontology of phenotypic qualities and multiple additional ontologies from the Open Biological Ontologies library. We also show how these logical definitions can be used for data integration when combined with a unified multi-species anatomy ontology.
Figures

References
-
- Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–290. - PubMed
-
- Khaja R, Zhang J, MacDonald JR, He Y, Joseph-George AM, Wei J, Rafiq MA, Qian C, Shago M, Pantano L, Aburatani H, Jones K, Redon R, Hurles M, Armengol L, Estivill X, Mural RJ, Lee C, Scherer SW, Feuk L. Genome assembly comparison identifies structural variants in the human genome. Nat Genet. 2006;38:1413–1418. - PMC - PubMed
-
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–1255. - PMC - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. - PMC - PubMed
-
- Mabee PM, Ashburner M, Cronk Q, Gkoutos GV, Haendel M, Segerdell E, Mungall C, Westerfield M. Phenotype ontologies: the bridge between genomics and evolution. Trends Ecol Evol. 2007;22:345–350. - PubMed
Publication types
MeSH terms
Grants and funding
- HG000330/HG/NHGRI NIH HHS/United States
- BG/G004358/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G8225539/MRC_/Medical Research Council/United Kingdom
- R01 HG004838/HG/NHGRI NIH HHS/United States
- BB/G004358/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

