Immunohistochemical analysis of oxidative stress and DNA repair proteins in normal mammary and breast cancer tissues
- PMID: 20064251
- PMCID: PMC2830938
- DOI: 10.1186/1471-2407-10-9
Immunohistochemical analysis of oxidative stress and DNA repair proteins in normal mammary and breast cancer tissues
Abstract
Background: During the course of normal cellular metabolism, oxygen is consumed and reactive oxygen species (ROS) are produced. If not effectively dissipated, ROS can accumulate and damage resident proteins, lipids, and DNA. Enzymes involved in redox regulation and DNA repair dissipate ROS and repair the resulting damage in order to preserve a functional cellular environment. Because increased ROS accumulation and/or unrepaired DNA damage can lead to initiation and progression of cancer and we had identified a number of oxidative stress and DNA repair proteins that influence estrogen responsiveness of MCF-7 breast cancer cells, it seemed possible that these proteins might be differentially expressed in normal mammary tissue, benign hyperplasia (BH), ductal carcinoma in situ (DCIS) and invasive breast cancer (IBC).
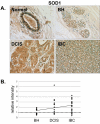
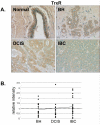
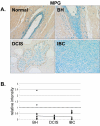
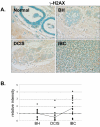
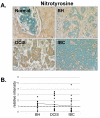
Methods: Immunohistochemistry was used to examine the expression of a number of oxidative stress proteins, DNA repair proteins, and damage markers in 60 human mammary tissues which were classified as BH, DCIS or IBC. The relative mean intensity was determined for each tissue section and ANOVA was used to detect statistical differences in the relative expression of BH, DCIS and IBC compared to normal mammary tissue.
Results: We found that a number of these proteins were overexpressed and that the cellular localization was altered in human breast cancer tissue.
Conclusions: Our studies suggest that oxidative stress and DNA repair proteins not only protect normal cells from the damaging effects of ROS, but may also promote survival of mammary tumor cells.
Figures







Similar articles
-
Progression-specific genes identified in microdissected formalin-fixed and paraffin-embedded tissue containing matched ductal carcinoma in situ and invasive ductal breast cancers.BMC Med Genomics. 2018 Sep 20;11(1):80. doi: 10.1186/s12920-018-0403-5. BMC Med Genomics. 2018. PMID: 30236106 Free PMC article.
-
Loss of interferon regulatory factor 5 (IRF5) expression in human ductal carcinoma correlates with disease stage and contributes to metastasis.Breast Cancer Res. 2011;13(6):R111. doi: 10.1186/bcr3053. Epub 2011 Nov 4. Breast Cancer Res. 2011. PMID: 22053985 Free PMC article.
-
Profiling differential microRNA expression between in situ, infiltrative and lympho-vascular space invasive breast cancer: a pilot study.Clin Exp Metastasis. 2018 Feb;35(1-2):3-13. doi: 10.1007/s10585-017-9868-4. Epub 2017 Dec 6. Clin Exp Metastasis. 2018. PMID: 29214365
-
Mammary stem cells and breast cancer--role of Notch signalling.Stem Cell Rev. 2007 Jun;3(2):169-75. doi: 10.1007/s12015-007-0023-5. Stem Cell Rev. 2007. PMID: 17873349 Review.
-
Redox environment, free radical, and oxidative DNA damage.Antioxid Redox Signal. 2013 Jun 20;18(18):2399-408. doi: 10.1089/ars.2012.4920. Epub 2013 Feb 6. Antioxid Redox Signal. 2013. PMID: 23249296 Review.
Cited by
-
Effect of higher frequency components and duration of vibration on bone tissue alterations in the rat-tail model.Ind Health. 2015;53(3):245-59. doi: 10.2486/indhealth.2014-0117. Epub 2015 Apr 4. Ind Health. 2015. PMID: 25843564 Free PMC article.
-
BRCA1 mutations drive oxidative stress and glycolysis in the tumor microenvironment: implications for breast cancer prevention with antioxidant therapies.Cell Cycle. 2012 Dec 1;11(23):4402-13. doi: 10.4161/cc.22776. Epub 2012 Nov 21. Cell Cycle. 2012. PMID: 23172369 Free PMC article.
-
Naturally occurring germline and tumor-associated mutations within the ATP-binding motifs of PTEN lead to oxidative damage of DNA associated with decreased nuclear p53.Hum Mol Genet. 2011 Jan 1;20(1):80-9. doi: 10.1093/hmg/ddq434. Epub 2010 Oct 6. Hum Mol Genet. 2011. PMID: 20926450 Free PMC article.
-
Chlorine, chromium, proteins of oxidative stress and DNA repair pathways are related to prognosis in oral cancer.Sci Rep. 2021 Nov 16;11(1):22314. doi: 10.1038/s41598-021-01753-x. Sci Rep. 2021. PMID: 34785721 Free PMC article.
-
Interplay between reactive oxygen species and ERK activation in cervical cancer cells.Front Cell Dev Biol. 2024 Nov 19;12:1465729. doi: 10.3389/fcell.2024.1465729. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39629272 Free PMC article.
References
-
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11(3):298–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

