Molecular and cellular evidence for biased mitotic gene conversion in hybrid scallop
- PMID: 20064268
- PMCID: PMC2818637
- DOI: 10.1186/1471-2148-10-6
Molecular and cellular evidence for biased mitotic gene conversion in hybrid scallop
Abstract
Background: Concerted evolution has been believed to account for homogenization of genes within multigene families. However, the exact mechanisms involved in the homogenization have been under debate. Use of interspecific hybrid system allows detection of greater level of sequence variation, and therefore, provide advantage for tracing the sequence changes. In this work, we have used an interspecific hybrid system of scallop to study the sequence homogenization processes of rRNA genes.
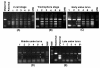
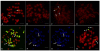
Results: Through the use of a hybrid scallop system (Chlamys farreri female symbol x Argopecten irradians male symbol), here we provide solid molecular and cellular evidence for homogenization of the rDNA sequences into maternal genotypes. The ITS regions of the rDNA of the two scallop species exhibit distinct sequences and thereby restriction fragment length polymorphism (RFLP) patterns, and such a difference was exploited to follow the parental ITS contributions in the F1 hybrid during early development using PCR-RFLP. The representation of the paternal ITS decreased gradually in the hybrid during the development of the hybrid, and almost diminished at the 14th day after fertilization while the representation of the maternal ITS gradually increased. Chromosomal-specific fluorescence in situ hybridization (FISH) analysis in the hybrid revealed the presence of maternal ITS sequences on the paternal ITS-bearing chromosomes, but not vice versa. Sequence analysis of the ITS region in the hybrid not only confirmed the maternally biased conversion, but also allowed the detection of six recombinant variants in the hybrid involving short recombination regions, suggesting that site-specific recombination may be involved in the maternally biased gene conversion.
Conclusion: Taken together, these molecular and cellular evidences support rapid concerted gene evolution via maternally biased gene conversion. As such a process would lead to the expression of only one parental genotype, and have the opportunities to generate recombinant intermediates; this work may also have implications in novel hybrid zone alleles and genetic imprinting, as well as in concerted gene evolution. In the course of evolution, many species may have evolved involving some levels of hybridization, intra- or interspecific, the sex-biased sequence homogenization could have led to a greater role of one sex than the other in some species.
Figures


References
-
- Hughes KW, Petersen RH. Apparent recombination or gene conversion in the ribosomal ITS region of a Flammulina (Fungi, Agaricales) hybrid. Mol Biol Evol. 2001;18(1):94–96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

