A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection
- PMID: 20064372
- PMCID: PMC2892837
- DOI: 10.1016/j.cell.2009.12.018
A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection
Abstract
During the course of a viral infection, viral proteins interact with an array of host proteins and pathways. Here, we present a systematic strategy to elucidate the dynamic interactions between H1N1 influenza and its human host. A combination of yeast two-hybrid analysis and genome-wide expression profiling implicated hundreds of human factors in mediating viral-host interactions. These factors were then examined functionally through depletion analyses in primary lung cells. The resulting data point to potential roles for some unanticipated host and viral proteins in viral infection and the host response, including a network of RNA-binding proteins, components of WNT signaling, and viral polymerase subunits. This multilayered approach provides a comprehensive and unbiased physical and regulatory model of influenza-host interactions and demonstrates a general strategy for uncovering complex host-pathogen relationships.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures

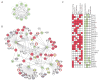


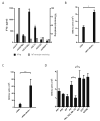
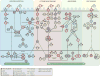
References
-
- Brander C, Walker BD. Modulation of host immune responses by clinically relevant human DNA and RNA viruses. Curr Opin Microbiol. 2000;3:379–386. - PubMed
-
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

