CtIP links DNA double-strand break sensing to resection
- PMID: 20064462
- PMCID: PMC2807415
- DOI: 10.1016/j.molcel.2009.12.002
CtIP links DNA double-strand break sensing to resection
Abstract
In response to DNA double-strand breaks (DSBs), cells sense the DNA lesions and then activate the protein kinase ATM. Subsequent DSB resection produces RPA-coated ssDNA that is essential for activation of the DNA damage checkpoint and DNA repair by homologous recombination (HR). However, the biochemical mechanism underlying the transition from DSB sensing to resection remains unclear. Using Xenopus egg extracts and human cells, we show that the tumor suppressor protein CtIP plays a critical role in this transition. We find that CtIP translocates to DSBs, a process dependent on the DSB sensor complex Mre11-Rad50-NBS1, the kinase activity of ATM, and a direct DNA-binding motif in CtIP, and then promotes DSB resection. Thus, CtIP facilitates the transition from DSB sensing to processing: it does so by binding to the DNA at DSBs after DSB sensing and ATM activation and then promoting DNA resection, leading to checkpoint activation and HR.
2009 Elsevier Inc.
Figures
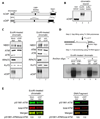
Schematic comparison of human CtIP and Xenopus CtIP (xCtIP).
xCtIP associated with damaged, but not undamaged, chromatin in the Xenopus extract, and chromatin-associated xCtIP was highly modified as evidenced by its reduced gel mobility. Xenopus extract (20 µl) was incubated with 10,000 EcoRI-treated or buffer-treated demembranated sperm nuclei/µl for 30 min. After incubation, chromatin was isolated and chromatin-associated xCtIP was detected by immunoblotting (lanes 2 and 3). Lane 1, xCtIP in 1 µl of total extract.
xCtIP is required for association of RPA and ATR, but not NBS1, ATM and Ku70, with chromatin containing DSBs. Left panel: EcoRI-treated sperm chromatin was incubated with mock-depleted or xCtIP-depleted extract for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting. xCtIP depletion blocked chromatin association of RPA70 and ATR, but not NBS1, ATM and Ku70. Right panel: Extract supplemented with 400 ng/µl of control IgG or affinity-purified, neutralizing xCtIP antibodies was incubated with EcoRI-treated sperm chromatin for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting. Addition of xCtIP antibodies inhibited chromatin association of xCtIP, RPA and ATR, but not NBS1, ATM and Ku70.
xCtIP is required for efficient DSB end resection in the Xenopus extract. Upper panel: A procedure for detecting DSB end resection in the Xenopus extract. The 3’ tail generated by 5’ strand resection of a DNA DSB in chromatin is ligated to the 5’ phosphorylated end of an anchor oligo using T4 RNA ligase I. After ligation, a second, complementary oligo is annealed to the anchor oligo and T4 DNA polymerase is used to fill the gap between the 3’ end of the second oligo and the 5’ resected end in chromatin in the presence of 32P-α-dCTP (see Experimental Procedures in Supplemental Information for details). The amount of 32P-α-dCTP incorporation represents the extent of DSB resection. Lower panels: EcoRI-treated sperm chromatin was incubated with mock-depleted (lane 2) or xCtIP-depleted extract (lane 3), or with extract supplemented with 400 ng/µl of control IgG (lane 5) or neutralizing xCtIP antibodies (lane 6) for 30 min. After incubation, chromatin was isolated and DSB resection was analyzed on agarose gels after oligo anchoring and gap filling in the presence of 32P-α-dCTP. The anchor oligo was omitted from the control lanes 1 and 4.
Immunodepletion of xCtIP from the Xenopus extract increased the extent of ATM autophosphorylation at S1981 induced by EcoRI-treated sperm chromatin or by dsDNA fragments, although xCtIP depletion partially removed ATM from the extract. Mock-depleted or xCtIP-depleted extract was incubated with 10,000 sperm nuclei/µl (left panel) or 5 ng/µl of a 2 kbp dsDNA fragment (right panel) for 25 min. After incubation, immunoblotting was performed to detect activated ATM and total ATM in the extracts with rabbit phospho-S1981 specific anti-ATM antibodies and guinea pig anti-Xenopus ATM antibodies, respectively. Primary antibodies were detected by Alexa Fluor 680-labeled anti-rabbit secondary antibodies and IRdye 800-labeled anti-guinea pig second antibodies using Odyssey infrared imaging system. Immunoblotting signals were quantified using Odyssey 3.0 software.
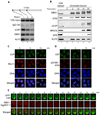
Association of xCtIP, NBS1 and Ku70 with different regions of a DNA fragment incubated in the Xenopus extract. A 7.4 kbp linearized plasmid (5 ng/µl) was incubated with the extract for 15 min. After incubation, ChIP experiments were performed to assess the association of proteins at three representative regions of the DNA fragment: 97–298 bp (region I), 1857–2060 (region II) and 3711–3920 (region III).
Time-course analysis of chromatin association of NBS1, ATM, xCtIP, RPA70, ATR and Ku70 in the Xenopus extract in response to DSBs. EcoRI-treated chromatin incubated in the Xenopus extract was isolated at the indicated times and associated proteins were detected by immunoblotting. Signal in lane 1 represents NBS1, ATM, RPA70, ATR or Ku70 in 0.5 µl of total extract, or xCtIP in 0.1 µl of total extract.
Accumulation of CtIP and Mre11 at DNA damage sites in human cells at different times after DNA damage. Laser irradiation was performed to generate DSBs in a line pattern in human U2OS cells. Subsequently, cells were fixed at the indicated times followed by indirect immunofluorescence to assess the recruitment of CtIP and Mre11 to DNA damage sites. DNA was stained with DAPI.
Same as depicted in panel C, except that the accumulation of CtIP and pS1981-ATM at DNA damage sites was assessed at the indicated time points after laser irradiation.
Laser irradiation was performed in a U2OS cell co-transfected with GFP-CtIP and NBS1-TagRFP-T followed by live cell imaging to assess the kinetics of damage recruitment of CtIP and NBS1 to a damage line.
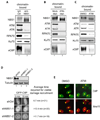
Addition of NBS1 neutralizing antibodies inhibited association of ATM, ATR, RPA70, Ku70 and xCtIP, but not NBS1 itself, with damaged chromatin in the Xenopus extract. Extract preincubated with 300 ng/µl of control IgG or affinity-purified NBS1 antibodies were incubated with EcoRI-treated sperm chromatin for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting.
ATM depletion abrogated association of xCtIP, RPA70 and ATR, but not NBS1 and Ku70, with damaged chromatin in the Xenopus extract. EcoRI-treated sperm chromatin was incubated with mock-depleted or ATM-depleted extract for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting.
ATM inhibition blocked the association of xCtIP, RPA70 and ATR, but not NBS1, Ku70 and ATM, with damaged chromatin in the Xenopus extract. EcoRI-treated sperm chromatin was incubated with the extract supplemented with DMSO or the ATM inhibitor KU-55933 (100 µM) for 30 min (ATMi). After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting.
Knockdown of NBS1 in human cells delayed damage recruitment of GFP-CtIP. Upper panel: Lentiviral vectors encoding a control non-targeting shRNA or NBS1-targeting shRNAs were transfected into cells and NBS1 protein levels were examined two days after transfection. Lower panel: GFP-CtIP and the shRNA-encoding vectors were co-transfected into U2OS cells. Two days after transfection, laser irradiation was performed on cells that were transfected with both GFP-CtIP and shRNA-encoding vectors (mCherry positive). Damage recruitment of GFP-CtIP to DNA damage sites was examined by live cell imaging for 20 min after laser irradiation. The average time required for detectable accumulation of GFP-CtIP at laser-induced DNA damage sites in cells treated with shCtrl, shNBS1-1 and shNBS1-2 is 6.7 min, >12.4 min and >11.7 min, respectively. Whereas GFP-CtIP exhibited damage recruitment in all shCtrl treated cells, no GFP-CtIP damage recruitment was observed in 25% of shNBS1-1 treated cells and 20% of shNBS1-2 treated cells 20 min after laser irradiation.
ATM inhibition resulted in delay in damage recruitment of CtIP, but not Mre11, in human cells. U2OS cells were treated with DMSO or the ATM inhibitor KU-55933 (10 µM) for 30 min. Subsequently, laser irradiation was performed to induce DSBs in cells. Ten min after irradiation cells were fixed followed by indirect immunofluorescence to assess the association of CtIP and Mre11 with DNA damage sites.
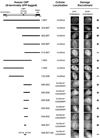
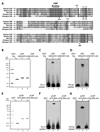
Sequence comparison of the DR motif and its surrounding regions in CtIP homologs in human, mouse, rat, chicken and Xenopus. The highly conserved CtBP-binding motif (“PLDLS”) is located immediately upstream of the DR motif (509–557) in human CtIP. Numbers above and below the sequences denote the amino acid positions in human CtIP and Xenopus CtIP, respectively.
Purified recombinant GST-His, GST-His tagged hCtIP(509–557) and GST-His tagged hCtIP(509–557)-AA proteins were resolved in a denaturing polyacrylamide gel and detected with Coomassie blue dye.
Recombinant proteins shown in panel B were incubated with a 5’ 32P-labled 83 nt ssDNA oligo for 10 min at room temperature prior to gel shift analysis. hCtIP(509–557), but not the GST-His tag or hCtIP(509–557)-AA, associated with the ssDNA oligo, resulting in a mobility shift of the DNA substrate on a native polyacrylamide gel.
Same as depicted for panel C, except that a 5’ 32P-labeled 100 bp dsDNA fragment was incubated with the recombinant proteins. hCtIP(509–557), but not the GST-His tag or hCtIP(509–557)-AA, associated with the dsDNA fragment.
Purified recombinant GST, GST-tagged xCtIP(479–584) and GST-tagged xCtIP(479–584)-AA proteins resolved in a denaturing polyacrylamide gel were detected with Coomassie blue dye.
Same as depicted for panel C except that recombinant proteins shown in panel E were used for gel mobility shift analysis. xCtIP(479–584), but not the GST tag or xCtIP(479–584)-AA, associated with the ssDNA oligo.
Same as depicted for panel D except that recombinant proteins shown in panel E were used for gel mobility shift analysis. xCtIP(479–584), but not the GST tag or xCtIP(479–584)-AA, associated with the dsDNA fragment.
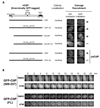
The DR motif is essential for damage recruitment of CtIP. N-terminally GFP-tagged, wild type or mutant CtIPs were transfected into U2OS cells or U2OS cells expressing an shRNA that depletes endogenous CtIP. Cells were then subjected to laser irradiation to induce DSBs. Damage recruitment of GFP-CtIP proteins was examined 20 min after laser irradiation.
Unlike GFP-CtIP, damage recruitment of GFP-CtIP(509–557) does not require ATM kinase activity. GFP-CtIP (containing full length CtIP) and GFP-CtIP(509–557) (containing the DR motif) were transfected into U2OS cells. Subsequently, cells were treated with DMSO or the ATM inhibitor KU-55933 (10 µM) for 30 min before laser irradiation. The damage recruitment kinetics of GFP-CtIP or GFP-CtIP(509–557) were assessed by live cell imaging.
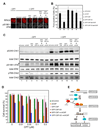
- A
and B. CtIP damage recruitment is required for efficient formation of RPA foci at DNA damage sites. Proliferating U2OS cells expressing a control shRNA (shControl), an shRNA targeting endogenous CtIP only (shCtIP), GFP-CtIP, GFP-CtIP-AA, GFP-CtIP and shCtIP or GFP-CtIP-AA and shCtIP were treated with 1 µM camptothecin (CPT) for 2 hr. Subsequently, cells were fixed and indirect immunofluorescence was performed using antibodies against RPA and pS1981-ATM. Primary antibodies were detected using donkey anti-mouse Alexa 568-labeled and donkey anti-rabbit Cy5-labled secondary antibodies. pS1981-ATM signal was used as a marker for CPT-induced DSB damage. Representative images were shown in A. The percentage of RPA foci-positive cells among the pS1981-ATM-positive cells for each sample is shown in B. Data represent the mean ± sd. Defects in RPA focus formation in CtIP-depleted cells were reversed by expression of GFP-CtIP, but not GFP-CtIP-AA.
- C
CtIP damage recruitment is required for efficient Chk1 phosphorylation after DSB damage. U2OS cells described in panel A were treated with 1 µM camptothecin for 2 hr followed by immunoblotting to assess Chk1 phosphorylation at S345, Chk2 phosphorylation at T68 and ATM autophosphorylation at S1981. Defects in Chk1 phosphorylation in CtIP-depleted cells were reversed by expression of GFP-CtIP, but not GFP-CtIP-AA.
- D
Mutations in the DR motif of CtIP caused hypersensitivity of cells to DSB damage. U2OS cells described in panel A were treated with various concentrations of camptothecin for 2 hr. Subsequently, 1000 treated cells were re-plated in triplicate and clonogenic analysis was performed to assess the sensitivity of the cells to DSB damage. Data represent mean ± sd. DNA damage hypersensitivity caused by CtIP depletion was reversed by expression of GFP-CtIP, but not GFP-CtIP-AA. Asterisk, nonspecific band.
- E
A model for damage recruitment of CtIP and its functional relationships with MRN and ATM in DSB resection. In response to DSBs, the DSB sensor complex MRN recruits ATM to the DNA/chromatin flanking a DNA end where ATM activation occurs. Following activation, ATM promotes recruitment of CtIP to DSBs, possibly by phosphorylating CtIP directly or indirectly. MRN may also directly facilitate CtIP damage recruitment. In addition to MRN and ATM, damage recruitment of CtIP requires its DR motif with direct DNA-binding activity. It is likely that conformational changes in CtIP after modifications (e.g. ATM-dependent phosphorylation and BRCA1-dependent ubiquitination) unmask the DR motif in CtIP, which then directly binds to the DNA at DSBs, leading to damage recruitment of CtIP. After damage recruitment, CtIP functions in cooperation with MRN to promote resection of 5’ strand DNA, resulting in generation of 3’ ssDNA tails that are bound by RPA. RPA-coated ssDNA then serves as a signal for activation of ATR and the DNA damage checkpoint. It also serves as a substrate for formation of the Rad51-ssDNA filament that is required for HR-mediated DSB repair.
References
-
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

