Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping
- PMID: 20064465
- PMCID: PMC2807993
- DOI: 10.1016/j.molcel.2009.12.003
Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping
Abstract
Recent transcriptome analysis indicates that > 90% of human genes undergo alternative splicing, underscoring the contribution of differential RNA processing to diverse proteomes in higher eukaryotic cells. The polypyrimidine tract-binding protein PTB is a well-characterized splicing repressor, but PTB knockdown causes both exon inclusion and skipping. Genome-wide mapping of PTB-RNA interactions and construction of a functional RNA map now reveal that dominant PTB binding near a competing constitutive splice site generally induces exon inclusion, whereas prevalent binding close to an alternative site often causes exon skipping. This positional effect was further demonstrated by disrupting or creating a PTB-binding site on minigene constructs and testing their responses to PTB knockdown or overexpression. These findings suggest a mechanism for PTB to modulate splice site competition to produce opposite functional consequences, which may be generally applicable to RNA-binding splicing factors to positively or negatively regulate alternative splicing in mammalian cells.
2009 Elsevier Inc.
Figures
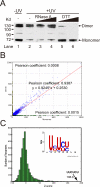
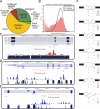


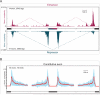

Comment in
-
RNA processing: Redrawing the map of charted territory.Mol Cell. 2009 Dec 25;36(6):918-9. doi: 10.1016/j.molcel.2009.12.004. Mol Cell. 2009. PMID: 20064456
References
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

