Decimal place slope, a fast and precise method for quantifying 13C incorporation levels for detecting the metabolic activity of microbial species
- PMID: 20064840
- PMCID: PMC2877982
- DOI: 10.1074/mcp.M900407-MCP200
Decimal place slope, a fast and precise method for quantifying 13C incorporation levels for detecting the metabolic activity of microbial species
Abstract
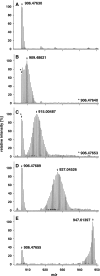
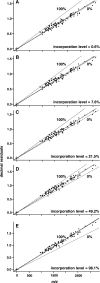
The metabolic incorporation of stable isotopes such as (13)C or (15)N into proteins has become a powerful tool for qualitative and quantitative proteome studies. We recently introduced a method that monitors heavy isotope incorporation into proteins and presented data revealing the metabolic activity of various species in a microbial consortium using this technique. To further develop our method using an liquid chromatography (LC)-mass spectrometry (MS)-based approach, we present here a novel approach for calculating the incorporation level of (13)C into peptides by using the information given in the decimal places of peptide masses obtained by modern high-resolution MS. In the present study, the applicability of this approach is demonstrated using Pseudomonas putida ML2 proteins uniformly labeled via the consumption of [(13)C(6)]benzene present in the medium at concentrations of 0, 10, 25, 50, and 100 atom %. The incorporation of (13)C was calculated on the basis of several labeled peptides derived from one band on an SDS-PAGE gel. The accuracy of the calculated incorporation level depended upon the number of peptide masses included in the analysis, and it was observed that at least 100 peptide masses were required to reduce the deviation below 4 atom %. This accuracy was comparable with calculations of incorporation based on the isotope envelope. Furthermore, this method can be extended to the calculation of the labeling efficiency for a wide range of biomolecules, including RNA and DNA. The technique will therefore allow a highly accurate determination of the carbon flux in microbial consortia with a direct approach based solely on LC-MS.
Figures
References
-
- Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem 389, 1017–1031 - PubMed
-
- Friedrich M. W. (2006) Stable-isotope probing of DNA: insights into the function of uncultivated microorganisms from isotopically labeled metagenomes. Curr. Opin. Biotech 17, 59–66 - PubMed
-
- Jehmlich N., Schmidt F., von Bergen M., Richnow H. H., Vogt C. (2008) Protein-based stable isotope probing (Protein-SIP) reveals active species within anoxic mixed cultures. ISME J 2, 1122–1133 - PubMed
-
- Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

