Increased origin activity in transformed versus normal cells: identification of novel protein players involved in DNA replication and cellular transformation
- PMID: 20064876
- PMCID: PMC2853114
- DOI: 10.1093/nar/gkp1192
Increased origin activity in transformed versus normal cells: identification of novel protein players involved in DNA replication and cellular transformation
Abstract
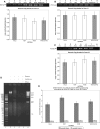
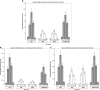
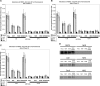
Using libraries of replication origins generated previously, we identified three clones that supported the autonomous replication of their respective plasmids in transformed, but not in normal cells. Assessment of their in vivo replication activity by in situ chromosomal DNA replication assays revealed that the chromosomal loci corresponding to these clones coincided with chromosomal replication origins in all cell lines, which were more active by 2-3-fold in the transformed by comparison to the normal cells. Evaluation of pre-replication complex (pre-RC) protein abundance at these origins in transformed and normal cells by chromatin immunoprecipitation assays, using anti-ORC2, -cdc6 and -cdt1 antibodies, showed that they were bound by these pre-RC proteins in all cell lines, but a 2-3-fold higher abundance was observed in the transformed by comparison to the normal cells. Electrophoretic mobility shift assays (EMSAs) performed on the most efficiently replicating clone, using nuclear extracts from the transformed and normal cells, revealed the presence of a DNA replication complex in transformed cells, which was barely detectable in normal cells. Subsequent supershift EMSAs suggested the presence of transformation-specific complexes. Mass spectrometric analysis of these complexes revealed potential new protein players involved in DNA replication that appear to correlate with cellular transformation.
Figures
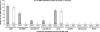



Similar articles
-
Comparative analysis of pre-replication complex proteins in transformed and normal cells.J Cell Biochem. 2012 Apr;113(4):1333-47. doi: 10.1002/jcb.24006. J Cell Biochem. 2012. PMID: 22134836
-
Two subunits of human ORC are dispensable for DNA replication and proliferation.Elife. 2016 Dec 1;5:e19084. doi: 10.7554/eLife.19084. Elife. 2016. PMID: 27906128 Free PMC article.
-
The human origin recognition complex is essential for pre-RC assembly, mitosis, and maintenance of nuclear structure.Elife. 2021 Feb 1;10:e61797. doi: 10.7554/eLife.61797. Elife. 2021. PMID: 33522487 Free PMC article.
-
Prospect of reprogramming replication licensing for cancer drug development.Biomed Pharmacother. 2021 Apr;136:111190. doi: 10.1016/j.biopha.2020.111190. Epub 2021 Jan 23. Biomed Pharmacother. 2021. PMID: 33497909 Review.
-
Cdt1 and geminin in DNA replication initiation.Subcell Biochem. 2012;62:71-87. doi: 10.1007/978-94-007-4572-8_5. Subcell Biochem. 2012. PMID: 22918581 Review.
Cited by
-
p53 gain-of-function mutations increase Cdc7-dependent replication initiation.EMBO Rep. 2017 Nov;18(11):2030-2050. doi: 10.15252/embr.201643347. Epub 2017 Sep 8. EMBO Rep. 2017. PMID: 28887320 Free PMC article.
-
Altered replication in human cells promotes DMPK (CTG)(n) · (CAG)(n) repeat instability.Mol Cell Biol. 2012 May;32(9):1618-32. doi: 10.1128/MCB.06727-11. Epub 2012 Feb 21. Mol Cell Biol. 2012. PMID: 22354993 Free PMC article.
-
Ku protein levels, localization and association to replication origins in different stages of breast tumor progression.J Cancer. 2013 May 26;4(5):358-70. doi: 10.7150/jca.6289. Print 2013. J Cancer. 2013. PMID: 23781282 Free PMC article.
-
Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins.J Cell Biol. 2011 Dec 26;195(7):1123-40. doi: 10.1083/jcb.201108121. J Cell Biol. 2011. PMID: 22201124 Free PMC article.
-
HBx protein of hepatitis B virus promotes reinitiation of DNA replication by regulating expression and intracellular stability of replication licensing factor CDC6.J Biol Chem. 2012 Jun 8;287(24):20545-54. doi: 10.1074/jbc.M112.359760. Epub 2012 Apr 19. J Biol Chem. 2012. PMID: 22523071 Free PMC article.
References
-
- Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 1963;28:329–348.
-
- Edenberg HJ, Huberman JA. Eukaryotic chromosome replication. Annu. Rev. Genet. 1975;9:245–284. - PubMed
-
- Hand R. Eucaryotic DNA: organization of the genome for replication. Cell. 1978;15:317–325. - PubMed
-
- Zannis-Hadjopoulos M, Price GB. Eukaryotic DNA replication. J. Cell. Biochem. 1999;Supplements 32/33:1–14. - PubMed
-
- Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. - PubMed

