Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA
- PMID: 20064924
- PMCID: PMC2838539
- DOI: 10.1093/hmg/ddq007
Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA
Abstract
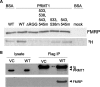
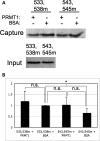
Fragile X syndrome is caused by the loss of expression of the fragile X mental retardation protein, FMRP. FMRP is an RNA-binding protein that is highly expressed in neurons and undergoes multiple post-translational modifications including methylation on arginine. FMRP is methylated on the high-affinity RNA-binding motif, the RGG box, at positions 533, 538, 543 and 545 of murine FMRP. To identify the arginines important for FMRP function, we examined their role in polyribosome and mRNA association. We found that arginines 533 and 538 were required for normal FMRP polyribosome association whereas all four arginines played a role in RNA binding, depending on the identity of the RNA. The model G-quadruplex RNA sc1 required arginines 533 and 538 for normal association with FMRP, whereas AATYK mRNA did not. In vitro methylation of FMRP-bearing arginine substitutions inhibited sc1 binding but not AATYK binding. In addition, we found that PRMT1 co-immunoprecipitated with FMRP isolated from cells and that siRNAs directed against PRMT1 led to reduced FMRP methylation. Thus, two lines of experimentation demonstrate that PRMT1 acts on FMRP in cells. In summary, we provide evidence for the important role of the RGG box in polyribosome association. We also demonstrate for the first time that the different arginines of the RGG box are important for the binding of different RNAs. Finally, we show that PRMT1 methylates FMRP in cells, suggesting a model where methylation of the RGG box modulates either the quantity or the identity of the RNAs bound by FMRP.
Figures


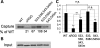


References
-
- Devys D., Lutz Y., Rouyer N., Bellocq J.-P., Mandel L. The FMR-1 protein is cytoplasmic, most abundant in neurons, and appears normal in carriers of the fragile X premutation. Nat. Genet. 1993;4:335–340. - PubMed
-
- Eberhart D.E., Malter H.E., Feng Y., Warren S.T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 1996;5:1083–1091. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

