Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells
- PMID: 20064925
- PMCID: PMC2838295
- DOI: 10.1074/jbc.M109.077081
Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells
Abstract
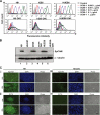
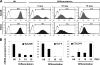
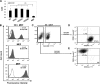
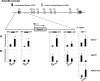
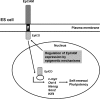
Human embryonic stem cells (hESCs) are unique pluripotent cells capable of self-renewal and differentiation into all three germ layers. To date, more cell surface markers capable of reliably identifying hESCs are needed. The epithelial cell adhesion molecule (EpCAM) is a type I transmembrane glycoprotein expressed in several progenitor cell populations and cancers. It has been used to enrich cells with tumor-initiating activity in xenograft transplantation studies. Here, we comprehensively profile the expression of EpCAM by immunofluorescence microscopy, Western blotting, and flow cytometry using an anti-EpCAM monoclonal antibody (mAb) OC98-1. We found EpCAM to be highly and selectively expressed by undifferentiated rather than differentiated hESCs. The protein and transcript level of EpCAM rapidly diminished as soon as hESC had differentiated. This silencing was closely and exclusively associated with the radical transformation of histone modification at the EpCAM promoter. Moreover, we demonstrated that the dynamic pattern of lysine 27 trimethylation of histone 3 was conferred by the interplay of SUZ12 and JMJD3, both of which were involved in maintaining hESC pluripotency. In addition, we used chromatin immunoprecipitation analysis to elucidate the direct regulation by EpCAM of several reprogramming genes, including c-MYC, OCT-4, NANOG, SOX2, and KLF4, to help maintain the undifferentiation of hESCs. Collectively, our results suggest that EpCAM might be used as a surface marker for hESC. The expression of EpCAM may be regulated by epigenetic mechanisms, and it is strongly associated with the maintenance of the undifferentiated state of hESCs.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

