Splicing of yeast aI5beta group I intron requires SUV3 to recycle MRS1 via mitochondrial degradosome-promoted decay of excised intron ribonucleoprotein (RNP)
- PMID: 20064926
- PMCID: PMC2838280
- DOI: 10.1074/jbc.M109.090761
Splicing of yeast aI5beta group I intron requires SUV3 to recycle MRS1 via mitochondrial degradosome-promoted decay of excised intron ribonucleoprotein (RNP)
Abstract
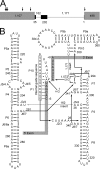
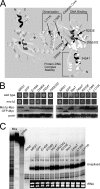
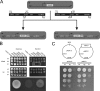
Yeast Suv3p is a member of the DEXH/D box family of RNA helicases and is a critical component of the mitochondrial degradosome, which also includes a 3' --> 5' exonuclease, Dss1p. Defects in the degradosome result in accumulation of aberrant transcripts, unprocessed transcripts, and excised group I introns. In addition, defects in SUV3 result in decreased splicing of the aI5beta and bI3 group I introns. Whereas a role for Suv3p in RNA degradation is well established, the function of Suv3p in splicing of group I introns has remained elusive. It has been particularly challenging to determine if Suv3p effects group I intron splicing through RNA degradation as part of the degradosome, or has a direct role in splicing as a chaperone, because nearly all perturbations of SUV3 or DSS1 result in loss of the mitochondrial genome. Here we utilized the suv3-1 allele, which is defective in RNA metabolism and yet maintains a stable mitochondrial genome, to investigate the role of Suv3p in splicing of the aI5beta group I intron. We provide genetic evidence that Mrs1p is a limiting cofactor for aI5beta splicing, and this evidence also suggests that Suv3p activity is required to recycle the excised aI5beta ribonucleoprotein. We also show that Suv3p acts indirectly as a component of the degradosome to promote aI5beta splicing. We present a model whereby defects in Suv3p result in accumulation of stable, excised group I intron ribonucleoproteins, which result in sequestration of Mrs1p, and a concomitant reduction in splicing of aI5beta.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

