Downregulation of PU.1 leads to decreased expression of Dectin-1 in alveolar macrophages during Pneumocystis pneumonia
- PMID: 20065023
- PMCID: PMC2825957
- DOI: 10.1128/IAI.01141-09
Downregulation of PU.1 leads to decreased expression of Dectin-1 in alveolar macrophages during Pneumocystis pneumonia
Abstract
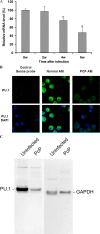
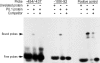
Dectin-1 is an important macrophage phagocytic receptor recognizing fungal beta-glucans. In this study, the mRNA levels of the Dectin-1 gene were found to be decreased by 61% in alveolar macrophages (AMs) from Pneumocystis-infected mice. The expression of Dectin-1 protein on the surface of these cells was also significantly decreased. By fluorescence in situ hybridization, mRNA expression levels of the transcription factor PU.1 were also found to be significantly reduced in AMs from Pneumocystis-infected mice. Electrophoretic mobility shift assay showed that PU.1 protein bound Dectin-1 gene promoter. With a luciferase reporter gene driven by the Dectin-1 gene promoter, the expression of the PU.1 gene in NIH 3T3 cells was found to enhance the luciferase activity in a dose-dependent manner. PU.1 expression knockdown by small interfering RNA (siRNA) caused a 63% decrease in Dectin-1 mRNA level and 40% decrease in protein level in AMs. Results of this study indicate that downregulation of PU.1 during Pneumocystis pneumonia leads to decreased expression of Dectin-1 in AMs.
Figures





References
-
- Bonfield, T. L., B. Raychaudhuri, A. Malur, S. Abraham, B. C. Trapnell, M. S. Kavuru, and M. J. Thomassen. 2003. PU.1 regulation of human alveolar macrophage differentiation requires granulocyte-macrophage colony-stimulating factor. Am. J. Physiol. Lung Cell. Mol. Physiol. 285:L1132-L1136. - PubMed
-
- Chang, H. C., S. Zhang, V. T. Thieu, R. B. Slee, H. A. Bruns, R. N. Laribee, M. J. Klemsz, and M. H. Kaplan. 2005. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity 22:693-703. - PubMed
-
- DeKoter, R. P., and H. Singh. 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288:1439-1441. - PubMed
-
- del Pilar Jimenez, A. M., S. Viriyakosol, L. Walls, S. K. Datta, T. Kirkland, S. E. Heinsbroek, G. Brown, and J. Fierer. 2008. Susceptibility to Coccidioides species in C57BL/6 mice is associated with expression of a truncated splice variant of Dectin-1 (Clec7a). Genes Immun. 9:338-348. - PMC - PubMed
-
- De Stefano, J. A., M. T. Cushion, R. G. Sleight, and P. D. Walzer. 1990. Analysis of Pneumocystis carinii cyst wall. I. Evidence for an outer surface membrane. J. Protozool. 37:428-435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

