Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11, and VirD4 are immunogenic components of a protective bacterial membrane vaccine
- PMID: 20065028
- PMCID: PMC2825951
- DOI: 10.1128/IAI.01207-09
Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11, and VirD4 are immunogenic components of a protective bacterial membrane vaccine
Abstract
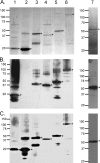
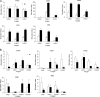
Anaplasma and related Ehrlichia spp. are important tick-borne, Gram-negative bacterial pathogens of livestock and humans that cause acute infection and disease and can persist. Immunization of cattle with an Anaplasma marginale fraction enriched in outer membranes (OM) can provide complete protection against disease and persistent infection. Serological responses of OM vaccinees to the OM proteome previously identified over 20 antigenic proteins, including three type IV secretion system (T4SS) proteins, VirB9-1, VirB9-2, and VirB10. Subsequent studies showed that these three proteins also stimulated CD4(+) T-cell responses in OM vaccinees. The T4SS, composed of a complex of proteins spanning the inner and outer membranes of certain bacteria, is an important virulence factor but is relatively unexplored as a vaccine target. The goal of this study was to determine if additional T4SS proteins are immunogenic for animals immunized with the protective OM fraction of A. marginale. T4SS proteins expressed by in vitro transcription and translation were screened for stimulating proliferation of T cells from OM vaccinees, and immunogenic proteins were expressed as recombinant proteins in Escherichia coli and their immunogenicity was verified. VirB2, a putative VirB7, VirB11, and VirD4 were immunogenic for OM vaccinees expressing several common major histocompatibility complex (MHC) class II haplotypes. VirB2 is encoded by multiple genes that share a conserved central region, and epitope mapping revealed T-cell epitopes in this region. The discovery of novel immunogenic T4SS proteins recognized by outbred individuals with common MHC haplotypes further justifies evaluating the T4SS as a potential vaccine candidate for pathogenic bacteria.
Figures



Similar articles
-
Association and evidence for linked recognition of type IV secretion system proteins VirB9-1, VirB9-2, and VirB10 in Anaplasma marginale.Infect Immun. 2012 Jan;80(1):215-27. doi: 10.1128/IAI.05798-11. Epub 2011 Oct 28. Infect Immun. 2012. PMID: 22038917 Free PMC article.
-
Breadth of the CD4+ T cell response to Anaplasma marginale VirB9-1, VirB9-2 and VirB10 and MHC class II DR and DQ restriction elements.Immunogenetics. 2012 Jul;64(7):507-23. doi: 10.1007/s00251-012-0606-4. Epub 2012 Feb 24. Immunogenetics. 2012. PMID: 22361828 Free PMC article.
-
Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine.Infect Immun. 2007 May;75(5):2333-42. doi: 10.1128/IAI.00061-07. Epub 2007 Mar 5. Infect Immun. 2007. PMID: 17339347 Free PMC article.
-
Persistent Infections and Immunity in Ruminants to Arthropod-Borne Bacteria in the Family Anaplasmataceae.Annu Rev Anim Biosci. 2016;4:177-97. doi: 10.1146/annurev-animal-022513-114206. Epub 2015 Dec 23. Annu Rev Anim Biosci. 2016. PMID: 26734888 Review.
-
Pseudomonas aeruginosa antigens as potential vaccines.FEMS Microbiol Rev. 1997 Nov;21(3):243-77. doi: 10.1111/j.1574-6976.1997.tb00353.x. FEMS Microbiol Rev. 1997. PMID: 9451816 Review.
Cited by
-
Phylogenomics reveals a diverse Rickettsiales type IV secretion system.Infect Immun. 2010 May;78(5):1809-23. doi: 10.1128/IAI.01384-09. Epub 2010 Feb 22. Infect Immun. 2010. PMID: 20176788 Free PMC article. Review.
-
PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species.Infect Immun. 2011 Nov;79(11):4286-98. doi: 10.1128/IAI.00207-11. Epub 2011 Sep 6. Infect Immun. 2011. PMID: 21896772 Free PMC article. Review.
-
Development of a Recombinase-Mediated Cassette Exchange System for Gene Knockout and Expression of Non-Native Gene Sequences in Rickettsia.Vaccines (Basel). 2025 Jan 22;13(2):109. doi: 10.3390/vaccines13020109. Vaccines (Basel). 2025. PMID: 40006656 Free PMC article.
-
CD4 T cell antigens from Staphylococcus aureus Newman strain identified following immunization with heat-killed bacteria.Clin Vaccine Immunol. 2012 Apr;19(4):477-89. doi: 10.1128/CVI.05642-11. Epub 2012 Feb 8. Clin Vaccine Immunol. 2012. PMID: 22323557 Free PMC article.
-
Metagenome diversity illuminates origins of pathogen effectors.bioRxiv [Preprint]. 2023 Feb 27:2023.02.26.530123. doi: 10.1101/2023.02.26.530123. bioRxiv. 2023. Update in: mBio. 2024 May 8;15(5):e0075923. doi: 10.1128/mbio.00759-23. PMID: 36909625 Free PMC article. Updated. Preprint.
References
-
- Abbott, J. R., G. H. Palmer, K. A. Kegerreis, P. F. Hetrick, C. J. Howard, J. C. Hope, and W. C. Brown. 2005. Rapid and long-term disappearance of CD4+ T lymphocyte responses specific for Anaplasma marginale major surface protein-2 (MSP2) in MSP2 vaccinates following challenge with live A. marginale. J. Immunol. 174:6702-6715. - PubMed
-
- Andrzejewska, J., S. K. Lee, P. Olbermann, N. Lotzing, E. Katzowitsch, B. Linz, M. Achtman, C. I. Kado, S. Suerbaum, and C. Josenhans. 2006. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J. Bacteriol. 188:5865-5877. - PMC - PubMed
-
- Araujo, F. R., C. M. Costa, C. A. Ramos, T. A. Farias, I. I. Souza, E. S. Melo, C. Elisei, G. M. Rosinha, C. O. Soares, S. P. Fragoso, and A. H. Fonseca. 2008. IgG and IgG2 antibodies from cattle naturally infected with Anaplasma marginale recognize the recombinant vaccine candidate antigens VirB9, VirB10, and elongation factor-Tu. Mem. Inst. Oswaldo Cruz 103:186-190. - PubMed
-
- Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

