Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation
- PMID: 20065033
- PMCID: PMC2832491
- DOI: 10.1128/MCB.01290-09
Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation
Abstract
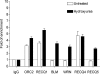
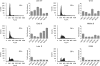
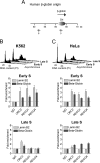
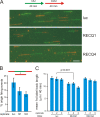
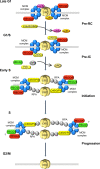
Cellular and biochemical studies support a role for all five human RecQ helicases in DNA replication; however, their specific functions during this process are unclear. Here we investigate the in vivo association of the five human RecQ helicases with three well-characterized human replication origins. We show that only RECQ1 (also called RECQL or RECQL1) and RECQ4 (also called RECQL4) associate with replication origins in a cell cycle-regulated fashion in unperturbed cells. RECQ4 is recruited to origins at late G(1), after ORC and MCM complex assembly, while RECQ1 and additional RECQ4 are loaded at origins at the onset of S phase, when licensed origins begin firing. Both proteins are lost from origins after DNA replication initiation, indicating either disassembly or tracking with the newly formed replisome. Nascent-origin DNA synthesis and the frequency of origin firing are reduced after RECQ1 depletion and, to a greater extent, after RECQ4 depletion. Depletion of RECQ1, though not that of RECQ4, also suppresses replication fork rates in otherwise unperturbed cells. These results indicate that RECQ1 and RECQ4 are integral components of the human replication complex and play distinct roles in DNA replication initiation and replication fork progression in vivo.
Figures
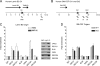



Similar articles
-
RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures.Cell Cycle. 2012 Nov 15;11(22):4252-65. doi: 10.4161/cc.22581. Epub 2012 Oct 24. Cell Cycle. 2012. PMID: 23095637 Free PMC article.
-
Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D.Mol Cancer. 2013 Apr 22;12(1):29. doi: 10.1186/1476-4598-12-29. Mol Cancer. 2013. PMID: 23601052 Free PMC article.
-
Interaction of RECQ4 and MCM10 is important for efficient DNA replication origin firing in human cells.Oncotarget. 2015 Dec 1;6(38):40464-79. doi: 10.18632/oncotarget.6342. Oncotarget. 2015. PMID: 26588054 Free PMC article.
-
Distinct roles of RECQ1 in the maintenance of genomic stability.DNA Repair (Amst). 2010 Mar 2;9(3):315-24. doi: 10.1016/j.dnarep.2009.12.010. Epub 2010 Jan 12. DNA Repair (Amst). 2010. PMID: 20061189 Free PMC article. Review.
-
Rothmund-Thomson syndrome helicase, RECQ4: on the crossroad between DNA replication and repair.DNA Repair (Amst). 2010 Mar 2;9(3):325-30. doi: 10.1016/j.dnarep.2010.01.006. Epub 2010 Jan 21. DNA Repair (Amst). 2010. PMID: 20096650 Review.
Cited by
-
Site-directed mutants of human RECQ1 reveal functional importance of the zinc binding domain.Mutat Res. 2016 Aug;790:8-18. doi: 10.1016/j.mrfmmm.2016.05.005. Epub 2016 May 17. Mutat Res. 2016. PMID: 27248010 Free PMC article.
-
A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells.Nat Commun. 2016 Jun 8;7:11748. doi: 10.1038/ncomms11748. Nat Commun. 2016. PMID: 27272143 Free PMC article.
-
"...Rewritten in the skin": clues to skin biology and aging from inherited disease.J Invest Dermatol. 2015 Jun;135(6):1484-1490. doi: 10.1038/jid.2015.88. Epub 2015 Mar 26. J Invest Dermatol. 2015. PMID: 25810110 Free PMC article. Review.
-
Trim33 masks a non-transcriptional function of E2f4 in replication fork progression.Nat Commun. 2023 Aug 23;14(1):5143. doi: 10.1038/s41467-023-40847-0. Nat Commun. 2023. PMID: 37612308 Free PMC article.
-
Replication Fork Remodeling and Therapy Escape in DNA Damage Response-Deficient Cancers.Front Oncol. 2020 May 5;10:670. doi: 10.3389/fonc.2020.00670. eCollection 2020. Front Oncol. 2020. PMID: 32432041 Free PMC article. Review.
References
-
- Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. - PubMed
-
- Bachrati, C. Z., and I. D. Hickson. 2008. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 117:219-233. - PubMed
-
- Bateman, J. R., and C. T. Wu. 2007. DNA replication and models for the origin of piRNAs. Bioessays 29:382-385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
