Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code
- PMID: 20065048
- PMCID: PMC2849384
- DOI: 10.1128/AAC.00906-09
Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code
Abstract
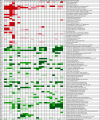
We have defined a sensitivity profile for 22 antibiotics by extending previous work testing the entire KEIO collection of close to 4,000 single-gene knockouts in Escherichia coli for increased susceptibility to 1 of 14 different antibiotics (ciprofloxacin, rifampin [rifampicin], vancomycin, ampicillin, sulfamethoxazole, gentamicin, metronidazole, streptomycin, fusidic acid, tetracycline, chloramphenicol, nitrofurantoin, erythromycin, and triclosan). We screened one or more subinhibitory concentrations of each antibiotic, generating more than 80,000 data points and allowing a reduction of the entire collection to a set of 283 strains that display significantly increased sensitivity to at least one of the antibiotics. We used this reduced set of strains to determine a profile for eight additional antibiotics (spectinomycin, cephradine, aztreonem, colistin, neomycin, enoxacin, tobramycin, and cefoxitin). The profiles for the 22 antibiotics represent a growing catalog of sensitivity fingerprints that can be separated into two components, multidrug-resistant mutants and those mutants that confer relatively specific sensitivity to the antibiotic or type of antibiotic tested. The latter group can be represented by a set of 20 to 60 strains that can be used for the rapid typing of antibiotics by generating a virtual bar code readout of the specific sensitivities. Taken together, these data reveal the complexity of intrinsic resistance and provide additional targets for the design of codrugs (or combinations of drugs) that potentiate existing antibiotics.
Figures


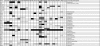


References
-
- Alekshun, M. N., and S. B. Levy.2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037-1050. - PubMed
-
- Allington, D. R., and M. P. Rivey.2001. Quinupristin/dalfopristin: a therapeutic review. Clin. Ther. 23:24-44. - PubMed
-
- Amundsen, S. K., and G. R. Smith.2003. Interchangeable parts of the Escherichia coli recombinational machinery. Cell 112:741-744. - PubMed
-
- Andrews, J. M.2001. Determination of minimum inhibitory concentrations. Antimicrob. Agents Chemother. 48:5-16. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

