Rescue of cardiomyopathy in peroxisome proliferator-activated receptor-alpha transgenic mice by deletion of lipoprotein lipase identifies sources of cardiac lipids and peroxisome proliferator-activated receptor-alpha activators
- PMID: 20065164
- PMCID: PMC2825753
- DOI: 10.1161/CIRCULATIONAHA.109.888735
Rescue of cardiomyopathy in peroxisome proliferator-activated receptor-alpha transgenic mice by deletion of lipoprotein lipase identifies sources of cardiac lipids and peroxisome proliferator-activated receptor-alpha activators
Abstract
Background: Emerging evidence in obesity and diabetes mellitus demonstrates that excessive myocardial fatty acid uptake and oxidation contribute to cardiac dysfunction. Transgenic mice with cardiac-specific overexpression of the fatty acid-activated nuclear receptor peroxisome proliferator-activated receptor-alpha (myosin heavy chain [MHC]-PPARalpha mice) exhibit phenotypic features of the diabetic heart, which are rescued by deletion of CD36, a fatty acid transporter, despite persistent activation of PPARalpha gene targets involved in fatty acid oxidation.
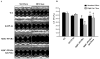
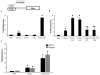
Methods and results: To further define the source of fatty acid that leads to cardiomyopathy associated with lipid excess, we crossed MHC-PPARalpha mice with mice deficient for cardiac lipoprotein lipase (hsLpLko). MHC-PPARalpha/hsLpLko mice exhibit improved cardiac function and reduced myocardial triglyceride content compared with MHC-PPARalpha mice. Surprisingly, in contrast to MHC-PPARalpha/CD36ko mice, the activity of the cardiac PPARalpha gene regulatory pathway is normalized in MHC-PPARalpha/hsLpLko mice, suggesting that PPARalpha ligand activity exists in the lipoprotein particle. Indeed, LpL mediated hydrolysis of very-low-density lipoprotein activated PPARalpha in cardiac myocytes in culture. The rescue of cardiac function in both models was associated with improved mitochondrial ultrastructure and reactivation of transcriptional regulators of mitochondrial function.
Conclusions: MHC-PPARalpha mouse hearts acquire excess lipoprotein-derived lipids. LpL deficiency rescues myocyte triglyceride accumulation, mitochondrial gene regulatory derangements, and contractile function in MHC-PPARalpha mice. Finally, LpL serves as a source of activating ligand for PPARalpha in the cardiomyocyte.
Figures





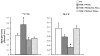
References
-
- Engelgau MM, Geiss LS, Saaddine JB, Boyle JP, Benjamin SM, Gregg EW, Tierney EF, Rios-Burrows N, Mokdad AH, Ford ES, Imperatore G, Narayan KM. The evolving diabetes burden in the United States. Ann Intern Med. 2004;140:945–950. - PubMed
-
- Zalesin KC, Franklin BA, Miller WM, Peterson ED, McCullough PA. Impact of obesity on cardiovascular disease. Endocrinol Metab Clin North Am. 2008;37:663–684. ix. - PubMed
-
- Abbott RD, Donahue RP, Kannel WB, Wilson PW. The impact of diabetes on survival following myocardial infarction in men vs. women. JAMA. 1988;260:3456–3460. - PubMed
-
- Cohen-Solal A, Beauvais F, Logeart D. Heart failure and diabetes mellitus: epidemiology and management of an alarming association. J Card Fail. 2008;14:615–625. - PubMed
-
- Cooper ME. Importance of advanced glycation end products in diabetes-associated cardiovascular and renal disease. Am J Hypertens. 2004;17:31S–38S. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL73029/HL/NHLBI NIH HHS/United States
- R01 HL045095/HL/NHLBI NIH HHS/United States
- 5P50HL07711304/HL/NHLBI NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
- P30 DK56341/DK/NIDDK NIH HHS/United States
- K12 HD001487/HD/NICHD NIH HHS/United States
- R37 HL045095/HL/NHLBI NIH HHS/United States
- P50 HL077113/HL/NHLBI NIH HHS/United States
- K08 HL08409/HL/NHLBI NIH HHS/United States
- R01 HL073029/HL/NHLBI NIH HHS/United States
- HL45095/HL/NHLBI NIH HHS/United States
- K12-HD001487/HD/NICHD NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- K08 HL084093/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

