A structural guide to proteins of the NF-kappaB signaling module
- PMID: 20066103
- PMCID: PMC2773632
- DOI: 10.1101/cshperspect.a000075
A structural guide to proteins of the NF-kappaB signaling module
Abstract
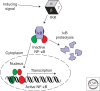
The prosurvival transcription factor NF-kappaB specifically binds promoter DNA to activate target gene expression. NF-kappaB is regulated through interactions with IkappaB inhibitor proteins. Active proteolysis of these IkappaB proteins is, in turn, under the control of the IkappaB kinase complex (IKK). Together, these three molecules form the NF-kappaB signaling module. Studies aimed at characterizing the molecular mechanisms of NF-kappaB, IkappaB, and IKK in terms of their three-dimensional structures have lead to a greater understanding of this vital transcription factor system.
Figures


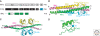
Similar articles
-
Regulation and function of IKK and IKK-related kinases.Sci STKE. 2006 Oct 17;2006(357):re13. doi: 10.1126/stke.3572006re13. Sci STKE. 2006. PMID: 17047224 Review.
-
A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity.Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. Epub 2007 May 8. Mol Syst Biol. 2007. PMID: 17486138 Free PMC article.
-
IkappaB kinase-independent IkappaBalpha degradation pathway: functional NF-kappaB activity and implications for cancer therapy.Mol Cell Biol. 2003 Nov;23(22):8070-83. doi: 10.1128/MCB.23.22.8070-8083.2003. Mol Cell Biol. 2003. PMID: 14585967 Free PMC article.
-
NF-kappaB p105 is a target of IkappaB kinases and controls signal induction of Bcl-3-p50 complexes.EMBO J. 1999 Sep 1;18(17):4766-78. doi: 10.1093/emboj/18.17.4766. EMBO J. 1999. PMID: 10469655 Free PMC article.
-
Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity.Annu Rev Immunol. 2000;18:621-63. doi: 10.1146/annurev.immunol.18.1.621. Annu Rev Immunol. 2000. PMID: 10837071 Review.
Cited by
-
A New Paradigm to Mitigate Osteosarcoma by Regulation of MicroRNAs and Suppression of the NF-κB Signaling Cascade.Dev Reprod. 2014 Dec;18(4):197-212. doi: 10.12717/devrep.2014.18.4.197. Dev Reprod. 2014. PMID: 25949190 Free PMC article.
-
Intrinsic protein disorder could be overlooked in cocrystallization conditions: An SRCD case study.Protein Sci. 2016 Nov;25(11):1977-1988. doi: 10.1002/pro.3010. Epub 2016 Aug 23. Protein Sci. 2016. PMID: 27508941 Free PMC article.
-
Role of nuclear factor κB in multiple sclerosis and experimental autoimmune encephalomyelitis.Neural Regen Res. 2018 Sep;13(9):1507-1515. doi: 10.4103/1673-5374.237109. Neural Regen Res. 2018. PMID: 30127103 Free PMC article. Review.
-
Folding and Stability of Ankyrin Repeats Control Biological Protein Function.Biomolecules. 2021 Jun 5;11(6):840. doi: 10.3390/biom11060840. Biomolecules. 2021. PMID: 34198779 Free PMC article. Review.
-
Selective regulation of a defined subset of inflammatory and immunoregulatory genes by an NF-κB p50-IκBζ pathway.Genes Dev. 2024 Jul 19;38(11-12):536-553. doi: 10.1101/gad.351630.124. Genes Dev. 2024. PMID: 38918046 Free PMC article.
References
-
- Agou F, Ye F, Goffinont S, Courtois G, Yamaoka S, Isräel A, Véron M 2002. NEMO trimerizes through its coiled-coiled C-terminal domain. J Biol Chem 277:17464–17475 - PubMed
-
- Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS 2003. A nucleosomal function for IκB kinase-α in NF-κB-dependent gene expression. Nature 423:659–663 - PubMed
-
- Baeuerle PA, Baltimore D 1988a. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-κB transcription factor. Cell 53:211–217 - PubMed
-
- Baeuerle PA, Baltimore D 1988b. IκB: A specific inhibitor of the NF-κB transcription factor. Science 242:540–546 - PubMed
-
- Bagnéris C, Ageichik AV, Cronin N, Wallace B, Collins M, Boshoff C, Waksman G, Barrett T 2008. Crystal structure of a vFlip-IKKγ complex: Insights into viral activation of the IKK signalosome. Mol Cell 30:620–631 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
