Prediction of functional class of proteins and peptides irrespective of sequence homology by support vector machines
- PMID: 20066123
- PMCID: PMC2789692
- DOI: 10.4137/bbi.s315
Prediction of functional class of proteins and peptides irrespective of sequence homology by support vector machines
Abstract
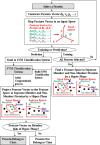
Various computational methods have been used for the prediction of protein and peptide function based on their sequences. A particular challenge is to derive functional properties from sequences that show low or no homology to proteins of known function. Recently, a machine learning method, support vector machines (SVM), have been explored for predicting functional class of proteins and peptides from amino acid sequence derived properties independent of sequence similarity, which have shown promising potential for a wide spectrum of protein and peptide classes including some of the low- and non-homologous proteins. This method can thus be explored as a potential tool to complement alignment-based, clustering-based, and structure-based methods for predicting protein function. This article reviews the strategies, current progresses, and underlying difficulties in using SVM for predicting the functional class of proteins. The relevant software and web-servers are described. The reported prediction performances in the application of these methods are also presented.
Keywords: machine learning method; peptide function; protein family; protein function; protein function prediction; support vector machines.
Figures

Similar articles
-
Homology-free prediction of functional class of proteins and peptides by support vector machines.Curr Protein Pept Sci. 2008 Feb;9(1):70-95. doi: 10.2174/138920308783565697. Curr Protein Pept Sci. 2008. PMID: 18336324 Review.
-
Recent progresses in the application of machine learning approach for predicting protein functional class independent of sequence similarity.Proteomics. 2006 Jul;6(14):4023-37. doi: 10.1002/pmic.200500938. Proteomics. 2006. PMID: 16791826 Review.
-
SVM-Fold: a tool for discriminative multi-class protein fold and superfamily recognition.BMC Bioinformatics. 2007 May 22;8 Suppl 4(Suppl 4):S2. doi: 10.1186/1471-2105-8-S4-S2. BMC Bioinformatics. 2007. PMID: 17570145 Free PMC article.
-
SVM-Prot 2016: A Web-Server for Machine Learning Prediction of Protein Functional Families from Sequence Irrespective of Similarity.PLoS One. 2016 Aug 15;11(8):e0155290. doi: 10.1371/journal.pone.0155290. eCollection 2016. PLoS One. 2016. PMID: 27525735 Free PMC article.
-
Neural network and SVM classifiers accurately predict lipid binding proteins, irrespective of sequence homology.J Theor Biol. 2014 Sep 7;356:213-22. doi: 10.1016/j.jtbi.2014.04.040. Epub 2014 May 10. J Theor Biol. 2014. PMID: 24819464
Cited by
-
Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently.Chem Soc Rev. 2015 Mar 7;44(5):1172-239. doi: 10.1039/c4cs00351a. Chem Soc Rev. 2015. PMID: 25503938 Free PMC article. Review.
-
Surveying alignment-free features for Ortholog detection in related yeast proteomes by using supervised big data classifiers.BMC Bioinformatics. 2018 May 3;19(1):166. doi: 10.1186/s12859-018-2148-8. BMC Bioinformatics. 2018. PMID: 29724166 Free PMC article.
-
Understanding the undelaying mechanism of HA-subtyping in the level of physic-chemical characteristics of protein.PLoS One. 2014 May 8;9(5):e96984. doi: 10.1371/journal.pone.0096984. eCollection 2014. PLoS One. 2014. PMID: 24809455 Free PMC article.
References
-
- Abbas AK, Lichtman AH.2005. Cellular and Molecular Immunology, Updated Edition. Saunders5th ed
-
- Aguilar D, Oliva B, Aviles FX, et al. TranScout: prediction of gene expression regulatory proteins from their sequences. Bioinformatics. 2002;18:597–607. - PubMed
-
- Al-Shahib A, Breitling R, Gilbert D. Feature selection and the class imbalance problem in predicting protein function from sequence. Appl Bioinformatics. 2005a;4:195–203. - PubMed
-
- Al-Shahib A, Breitling R, Gilbert D. FrankSum: new feature selection method for protein function prediction. Int. J. Neural Syst. 2005b;15:259–75. - PubMed
-
- Alexander S, Peters J, Mead A, et al. TiPS receptor and ion channel nomenclature supplement. Trends Pharmacol. Sci. 1999;19:5–85.
LinkOut - more resources
Full Text Sources

