Reversibly bound chloride in the atrial natriuretic peptide receptor hormone-binding domain: possible allosteric regulation and a conserved structural motif for the chloride-binding site
- PMID: 20066666
- PMCID: PMC2866279
- DOI: 10.1002/pro.332
Reversibly bound chloride in the atrial natriuretic peptide receptor hormone-binding domain: possible allosteric regulation and a conserved structural motif for the chloride-binding site
Abstract
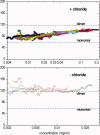
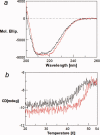
The binding of atrial natriuretic peptide (ANP) to its receptor requires chloride, and it is chloride concentration dependent. The extracellular domain (ECD) of the ANP receptor (ANPR) contains a chloride near the ANP-binding site, suggesting a possible regulatory role. The bound chloride, however, is completely buried in the polypeptide fold, and its functional role has remained unclear. Here, we have confirmed that chloride is necessary for ANP binding to the recombinant ECD or the full-length ANPR expressed in CHO cells. ECD without chloride (ECD(-)) did not bind ANP. Its binding activity was fully restored by bromide or chloride addition. A new X-ray structure of the bromide-bound ECD is essentially identical to that of the chloride-bound ECD. Furthermore, bromide atoms are localized at the same positions as chloride atoms both in the apo and in the ANP-bound structures, indicating exchangeable and reversible halide binding. Far-UV CD and thermal unfolding data show that ECD(-) largely retains the native structure. Sedimentation equilibrium in the absence of chloride shows that ECD(-) forms a strongly associated dimer, possibly preventing the structural rearrangement of the two monomers that is necessary for ANP binding. The primary and tertiary structures of the chloride-binding site in ANPR are highly conserved among receptor-guanylate cyclases and metabotropic glutamate receptors. The chloride-dependent ANP binding, reversible chloride binding, and the highly conserved chloride-binding site motif suggest a regulatory role for the receptor bound chloride. Chloride-dependent regulation of ANPR may operate in the kidney, modulating ANP-induced natriuresis.
Figures
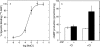
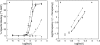
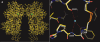


References
-
- de Bold AJ, Borenstein HB, Veress AT, Sonnenberg H. A rapid and potent natriuretic response to intravenous injection of atrial myocardial extract in rats. Life Science. 1981;28:89–94. - PubMed
-
- Grammer RT, Fukumi H, Inagami T, Misono KS. Rat atrial natriuretic factor. Purification and vasorelaxant activity. Biochem Biophys Res Commun. 1983;116:696–703. - PubMed
-
- Currie MG, Geller DM, Cole BR, Boylan JG, YuSheng W, Holmberg SW, Needleman P. Bioactive cardiac substances: potent vasorelaxant activity in mammalian atria. Science. 1983;221:71–73. - PubMed
-
- Ogawa H, Qiu Y, Ogata CM, Misono KS. Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domarotation mechanism for transmembrane signal transduction. J Biol Chem. 2004;279:28625–28631. - PubMed
-
- Misono KS, Ogawa H, Qiu Y, Ogata CM. Structural studies of the natriuretic peptide receptor: a novel hormone-induced rotation mechanism for transmembrane signal transduction. Peptides. 2005;26:957–968. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

