Incorporation of the fluorescent ribonucleotide analogue tCTP by T7 RNA polymerase
- PMID: 20067253
- PMCID: PMC2819109
- DOI: 10.1021/ac902456n
Incorporation of the fluorescent ribonucleotide analogue tCTP by T7 RNA polymerase
Abstract
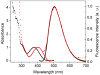
Fluorescent RNA is an important analytical tool in medical diagnostics, RNA cytochemistry, and RNA aptamer development. We have synthesized the fluorescent ribonucleotide analogue 1,3-diaza-2-oxophenothiazine-ribose-5'-triphosphate (tCTP) and tested it as substrate for T7 RNA polymerase in transcription reactions, a convenient route for generating RNA in vitro. When transcribing a guanine, T7 RNA polymerase incorporates tCTP with 2-fold higher catalytic efficiency than CTP and efficiently polymerizes additional NTPs onto the tC. Remarkably, T7 RNA polymerase does not incorporate tCTP with the same ambivalence opposite guanine and adenine with which DNA polymerases incorporate the analogous dtCTP. While several DNA polymerases discriminated against a d(tC-A) base pair only by factors <10, T7 RNA polymerase discriminates against tC-A base pair formation by factors of 40 and 300 when operating in the elongation and initiation mode, respectively. These catalytic properties make T7 RNA polymerase an ideal tool for synthesizing large fluorescent RNA, as we demonstrated by generating a approximately 800 nucleotide RNA in which every cytosine was replaced with tC.
Figures





Similar articles
-
A direct real-time spectroscopic investigation of the mechanism of open complex formation by T7 RNA polymerase.Biochemistry. 1996 Dec 10;35(49):15715-25. doi: 10.1021/bi960729d. Biochemistry. 1996. PMID: 8961934
-
Site-specific incorporation of fluorescent probes into RNA by specific transcription using unnatural base pairs.Nucleic Acids Symp Ser (Oxf). 2005;(49):287-8. doi: 10.1093/nass/49.1.287. Nucleic Acids Symp Ser (Oxf). 2005. PMID: 17150746
-
Enzymatic incorporation of emissive pyrimidine ribonucleotides.Chem Asian J. 2009 Mar 2;4(3):419-27. doi: 10.1002/asia.200800370. Chem Asian J. 2009. PMID: 19072942 Free PMC article.
-
Recent studies of T7 RNA polymerase mechanism.FEBS Lett. 1998 Dec 4;440(3):264-7. doi: 10.1016/s0014-5793(98)01484-7. FEBS Lett. 1998. PMID: 9872383 Review.
-
The structural changes of T7 RNA polymerase from transcription initiation to elongation.Curr Opin Struct Biol. 2009 Dec;19(6):683-90. doi: 10.1016/j.sbi.2009.09.001. Epub 2009 Oct 5. Curr Opin Struct Biol. 2009. PMID: 19811903 Free PMC article. Review.
Cited by
-
Live-Cell RNA Imaging with Metabolically Incorporated Fluorescent Nucleosides.J Am Chem Soc. 2022 Aug 17;144(32):14647-14656. doi: 10.1021/jacs.2c04142. Epub 2022 Aug 5. J Am Chem Soc. 2022. PMID: 35930766 Free PMC article.
-
Fluorescence Turn-On Sensing of DNA Duplex Formation by a Tricyclic Cytidine Analogue.J Am Chem Soc. 2017 Feb 1;139(4):1372-1375. doi: 10.1021/jacs.6b10410. Epub 2017 Jan 20. J Am Chem Soc. 2017. PMID: 28080035 Free PMC article.
-
Characterization of photophysical and base-mimicking properties of a novel fluorescent adenine analogue in DNA.Nucleic Acids Res. 2011 May;39(10):4513-24. doi: 10.1093/nar/gkr010. Epub 2011 Jan 29. Nucleic Acids Res. 2011. PMID: 21278417 Free PMC article.
-
Strategic base modifications refine RNA function and reduce CRISPR-Cas9 off-targets.Nucleic Acids Res. 2025 Feb 8;53(4):gkaf082. doi: 10.1093/nar/gkaf082. Nucleic Acids Res. 2025. PMID: 39964477 Free PMC article.
-
Synthesis and evaluation of an alkyne-modified ATP analog for enzymatic incorporation into RNA.Bioorg Med Chem Lett. 2016 Apr 1;26(7):1799-802. doi: 10.1016/j.bmcl.2016.02.038. Epub 2016 Feb 18. Bioorg Med Chem Lett. 2016. PMID: 26927424 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

