Integrating prokaryotes and eukaryotes: DNA transposases in light of structure
- PMID: 20067338
- PMCID: PMC3107681
- DOI: 10.3109/10409230903505596
Integrating prokaryotes and eukaryotes: DNA transposases in light of structure
Abstract
DNA rearrangements are important in genome function and evolution. Genetic material can be rearranged inadvertently during processes such as DNA repair, or can be moved in a controlled manner by enzymes specifically dedicated to the task. DNA transposases comprise one class of such enzymes. These move DNA segments known as transposons to new locations, without the need for sequence homology between transposon and target site. Several biochemically distinct pathways have evolved for DNA transposition, and genetic and biochemical studies have provided valuable insights into many of these. However, structural information on transposases - particularly with DNA substrates - has proven elusive in most cases. On the other hand, large-scale genome sequencing projects have led to an explosion in the number of annotated prokaryotic and eukaryotic mobile elements. Here, we briefly review biochemical and mechanistic aspects of DNA transposition, and propose that integrating sequence information with structural information using bioinformatics tools such as secondary structure prediction and protein threading can lead not only to an additional level of understanding but possibly also to testable hypotheses regarding transposition mechanisms. Detailed understanding of transposition pathways is a prerequisite for the long-term goal of exploiting DNA transposons as genetic tools and as a basis for genetic medical applications.
Conflict of interest statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Figures

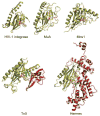
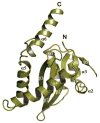
References
-
- Ariyoshi M, Vassylyev DG, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Atomic structure of the RuvC resolvase: A Holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
