Molecular modeling of the reductase domain to elucidate the reaction mechanism of reduction of peptidyl thioester into its corresponding alcohol in non-ribosomal peptide synthetases
- PMID: 20067617
- PMCID: PMC2835699
- DOI: 10.1186/1472-6807-10-1
Molecular modeling of the reductase domain to elucidate the reaction mechanism of reduction of peptidyl thioester into its corresponding alcohol in non-ribosomal peptide synthetases
Abstract
Background: Nonribosomal peptide synthetases (NRPSs) are multienzymatic, multidomain megasynthases involved in the biosynthesis of pharmaceutically important nonribosomal peptides. The peptaibol synthetase from Trichoderma virens (TPS) is an important member of the NRPS family that exhibits antifungal properties. The majority of the NRPSs terminate peptide synthesis with the thioesterase (TE) domain, which either hydrolyzes the thioester linkage, releasing the free peptic acid, or catalyzes the intramolecular macrocyclization to produce a macrolactone product. TPS is an important NRPS that does not encompass a TE domain, but rather a reductase domain (R domain) to release the mature peptide product reductively with the aid of a NADPH cofactor. However, the catalytic mechanism of the reductase domain has not yet been elucidated.
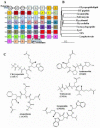
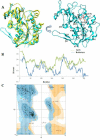
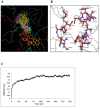
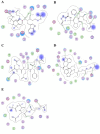
Results: We present here a three-dimensional (3D) model of the reductase domain based on the crystal structure of vestitone reductase (VR). VR belongs to the short-chain dehydrogenase/reductase (SDR) superfamily and is responsible for the nicotinamide dinucleotide phosphate (NADPH)-dependent reduction of the substrate into its corresponding secondary alcohol product. The binding sites of the probable linear substrates, alamethicin, trichotoxin, antiamoebin I, chrysopermin C and gramicidin, were identified within the modeled R domain using multiple docking approaches. The docking results of the ligand in the active site of the R domain showed that reductase side chains have a high affinity towards ligand binding, while the thioester oxygen of each substrate forms a hydrogen bond with the OH group of Tyr176 and the thiol group of the substrate is closer to the Glu220. The modeling and docking studies revealed the reaction mechanism of reduction of thioester into a primary alcohol.
Conclusion: Peptaibol biosynthesis incorporates a single R domain, which appears to catalyze the four-electron reduction reaction of a peptidyl carrier protein (PCP)-bound peptide to its corresponding primary alcohol. Analysis of R domains present in the non-redundant (nr) database of the NCBI showed that the R domain always resides in the last NRPS module and is involved in either a two or four-electron reduction reaction.
Figures

Similar articles
-
Structural insights into the regulation of NADPH binding to reductase domains of nonribosomal peptide synthetases: A concerted loop movement model.J Struct Biol. 2016 Jun;194(3):368-74. doi: 10.1016/j.jsb.2016.03.014. Epub 2016 Mar 16. J Struct Biol. 2016. PMID: 26993465
-
Structural characterization of a PCP-R didomain from an archaeal nonribosomal peptide synthetase reveals novel interdomain interactions.J Biol Chem. 2021 Jan-Jun;296:100432. doi: 10.1016/j.jbc.2021.100432. Epub 2021 Feb 18. J Biol Chem. 2021. PMID: 33610550 Free PMC article.
-
Nonprocessive [2 + 2]e- off-loading reductase domains from mycobacterial nonribosomal peptide synthetases.Proc Natl Acad Sci U S A. 2012 Apr 10;109(15):5681-6. doi: 10.1073/pnas.1118680109. Epub 2012 Mar 26. Proc Natl Acad Sci U S A. 2012. PMID: 22451903 Free PMC article.
-
New Structural Data Reveal the Motion of Carrier Proteins in Nonribosomal Peptide Synthesis.Angew Chem Int Ed Engl. 2016 Aug 16;55(34):9834-40. doi: 10.1002/anie.201602614. Epub 2016 Jul 20. Angew Chem Int Ed Engl. 2016. PMID: 27435901 Free PMC article. Review.
-
Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: discovery, dissection and diversity.Biochim Biophys Acta Proteins Proteom. 2017 Nov;1865(11 Pt B):1587-1604. doi: 10.1016/j.bbapap.2017.05.010. Epub 2017 May 16. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 28526268 Review.
Cited by
-
Engineering carboxylic acid reductase for selective synthesis of medium-chain fatty alcohols in yeast.Proc Natl Acad Sci U S A. 2020 Sep 15;117(37):22974-22983. doi: 10.1073/pnas.2010521117. Epub 2020 Sep 1. Proc Natl Acad Sci U S A. 2020. PMID: 32873649 Free PMC article.
-
MLACP: machine-learning-based prediction of anticancer peptides.Oncotarget. 2017 Aug 19;8(44):77121-77136. doi: 10.18632/oncotarget.20365. eCollection 2017 Sep 29. Oncotarget. 2017. PMID: 29100375 Free PMC article.
-
Biosynthesis of depsipeptides, or Depsi: The peptides with varied generations.Protein Sci. 2020 Dec;29(12):2316-2347. doi: 10.1002/pro.3979. Epub 2020 Nov 2. Protein Sci. 2020. PMID: 33073901 Free PMC article. Review.
-
AIPpred: Sequence-Based Prediction of Anti-inflammatory Peptides Using Random Forest.Front Pharmacol. 2018 Mar 27;9:276. doi: 10.3389/fphar.2018.00276. eCollection 2018. Front Pharmacol. 2018. PMID: 29636690 Free PMC article.
-
Distribution of amino acids in functional sites of proteins with high melting temperature.Bioinformation. 2012;8(23):1176-81. doi: 10.6026/97320630081176. Epub 2012 Nov 23. Bioinformation. 2012. PMID: 23275716 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

