Differential gene expression in incompatible interaction between wheat and stripe rust fungus revealed by cDNA-AFLP and comparison to compatible interaction
- PMID: 20067621
- PMCID: PMC2817678
- DOI: 10.1186/1471-2229-10-9
Differential gene expression in incompatible interaction between wheat and stripe rust fungus revealed by cDNA-AFLP and comparison to compatible interaction
Abstract
Background: Stripe rust of wheat, caused by Puccinia striiformis f. sp. tritici (Pst), is one of the most important diseases of wheat worldwide. Due to special features of hexaploid wheat with large and complex genome and difficulties for transformation, and of Pst without sexual reproduction and hard to culture on media, the use of most genetic and molecular techniques in studying genes involved in the wheat-Pst interactions has been largely limited. The objective of this study was to identify transcriptionally regulated genes during an incompatible interaction between wheat and Pst using cDNA-AFLP technique
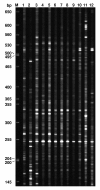
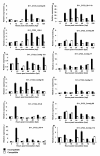
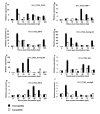
Results: A total of 52,992 transcript derived fragments (TDFs) were generated with 64 primer pairs and 2,437 (4.6%) of them displayed altered expression patterns after inoculation with 1,787 up-regulated and 650 down-regulated. We obtained reliable sequences (>100 bp) for 255 selected TDFs, of which 113 (44.3%) had putative functions identified. A large group (17.6%) of these genes shared high homology with genes involved in metabolism and photosynthesis; 13.8% to genes with functions related to disease defense and signal transduction; and those in the remaining groups (12.9%) to genes involved in transcription, transport processes, protein metabolism, and cell structure, respectively. Through comparing TDFs identified in the present study for incompatible interaction and those identified in the previous study for compatible interactions, 161 TDFs were shared by both interactions, 94 were expressed specifically in the incompatible interaction, of which the specificity of 43 selected transcripts were determined using quantitative real-time polymerase chain reaction (qRT-PCR). Based on the analyses of homology to genes known to play a role in defense, signal transduction and protein metabolism, 20 TDFs were chosen and their expression patterns revealed by the cDNA-AFLP technique were confirmed using the qRT-PCR analysis.
Conclusion: We uncovered a number of new candidate genes possibly involved in the interactions of wheat and Pst, of which 11 TDFs expressed specifically in the incompatible interaction. Resistance to stripe rust in wheat cv. Suwon11 is executed after penetration has occurred. Moreover, we also found that plant responses in compatible and incompatible interactions are qualitatively similar but quantitatively different soon after stripe rust fungus infection.
Figures


Similar articles
-
Stage-specific reprogramming of gene expression characterizes Lr48-mediated adult plant leaf rust resistance in wheat.Funct Integr Genomics. 2015 Mar;15(2):233-45. doi: 10.1007/s10142-014-0416-x. Epub 2014 Nov 29. Funct Integr Genomics. 2015. PMID: 25432546
-
cDNA-AFLP analysis reveals differential gene expression in compatible interaction of wheat challenged with Puccinia striiformis f. sp. tritici.BMC Genomics. 2009 Jun 30;10:289. doi: 10.1186/1471-2164-10-289. BMC Genomics. 2009. PMID: 19566949 Free PMC article.
-
Identification of expressed genes during compatible interaction between stripe rust (Puccinia striiformis) and wheat using a cDNA library.BMC Genomics. 2009 Dec 8;10:586. doi: 10.1186/1471-2164-10-586. BMC Genomics. 2009. PMID: 19995415 Free PMC article.
-
Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew.BMC Genomics. 2014 Oct 15;15(1):898. doi: 10.1186/1471-2164-15-898. BMC Genomics. 2014. PMID: 25318379 Free PMC article.
-
Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici.Mol Plant Pathol. 2014 Jun;15(5):433-46. doi: 10.1111/mpp.12116. Mol Plant Pathol. 2014. PMID: 24373199 Free PMC article. Review.
Cited by
-
Identification of two putative reference genes from grapevine suitable for gene expression analysis in berry and related tissues derived from RNA-Seq data.BMC Genomics. 2013 Dec 13;14:878. doi: 10.1186/1471-2164-14-878. BMC Genomics. 2013. PMID: 24330674 Free PMC article.
-
Potato calcium sensor modules StCBL3-StCIPK7 and StCBL3-StCIPK24 negatively regulate plant immunity.BMC Plant Biol. 2024 Jan 5;24(1):30. doi: 10.1186/s12870-023-04713-x. BMC Plant Biol. 2024. PMID: 38182981 Free PMC article.
-
Identification of reference genes suitable for qRT-PCR in grapevine and application for the study of the expression of genes involved in pterostilbene synthesis.Mol Genet Genomics. 2011 Apr;285(4):273-85. doi: 10.1007/s00438-011-0607-2. Epub 2011 Feb 22. Mol Genet Genomics. 2011. PMID: 21340517
-
Gene expression profiling by cDNA-AFLP reveals potential candidate genes for partial resistance of 'Président Roulin' against Venturia inaequalis.BMC Genomics. 2014 Nov 29;15:1043. doi: 10.1186/1471-2164-15-1043. BMC Genomics. 2014. PMID: 25433532 Free PMC article.
-
Stage-specific reprogramming of gene expression characterizes Lr48-mediated adult plant leaf rust resistance in wheat.Funct Integr Genomics. 2015 Mar;15(2):233-45. doi: 10.1007/s10142-014-0416-x. Epub 2014 Nov 29. Funct Integr Genomics. 2015. PMID: 25432546
References
-
- Yang YN, Shah J, Klessig DF. Signal perception and transduction in plant defense responses. Genes & Development. 1997;11:1621–1639. - PubMed
-
- Ebel J, Cosio EG. Elicitors of plant defense responses. International review of cytology. 1994;148:1–36. doi: 10.1016/S0074-7696(08)62404-3. - DOI
-
- Dixon RA, Harrison MJ, Lamb CJ. Early events in the activation of plant defense responses. Annual Review of Phytopathology. 1994;32:479–501. doi: 10.1146/annurev.py.32.090194.002403. - DOI
-
- Zhu Q, Dröge-Laser W, Dixon RA, Lamb C. Transcriptional activation of plant defense genes. Current Opinion in Genetics & Development. 1996;6:624–630. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

