Genetic analysis of the Staphylococcus epidermidis macromolecular synthesis operon: Serp1129 is an ATP binding protein and sigA transcription is regulated by both sigma(A)- and sigma(B)-dependent promoters
- PMID: 20067631
- PMCID: PMC2824700
- DOI: 10.1186/1471-2180-10-8
Genetic analysis of the Staphylococcus epidermidis macromolecular synthesis operon: Serp1129 is an ATP binding protein and sigA transcription is regulated by both sigma(A)- and sigma(B)-dependent promoters
Abstract
Background: The highly conserved macromolecular synthesis operon (MMSO) contains both dnaG (primase) and sigA (primary sigma factor). However, in previously evaluated gram-positive species, the MMSO is divergent upstream of dnaG. The MMSO of Bacillus subtilis contains three open reading frames (ORFs) that are differentially regulated by multiple promoters. In conjunction with studies to determine the expression profile of dnaG, the MMSO of Staphylococus epidermidis was characterized.
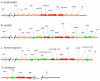
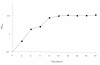
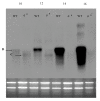
Results: The ORFs of S. epidermidis were compared to the previously described MMSO of B. subtilis and two additional ORFs in S. epidermidis, serp1129 and serp1130, were identified. The largest transcript, 4.8 kb in length, was expressed only in exponential growth and encompassed all four ORFs (serp1130, serp1129, dnaG, and sigA). A separate transcript (1.5 kb) comprising serp1130 and serp1129 was expressed in early exponential growth. Two smaller transcripts 1.3 and 1.2 kb in size were detected with a sigA probe in both exponential and post-exponential phases of growth. Western blot analysis correlated with the transcriptional profile and demonstrated that Serp1129 was detected only in the exponential phase of growth. Computational analysis identified that Serp1130 contained a CBS motif whereas Serp1129 contained an ATP/GTP binding motif. Functional studies of Serp1129 demonstrated that it was capable of binding both ATP and GTP. Comparisons with a sigB:dhfr mutant revealed that the 1.3 kb sigA transcript was regulated by a sigma(B)-dependent promoter.
Conclusions: These studies demonstrated that the S. epidermidis 1457 MMSO contains two ORFs (serp1129 and serp1130) not described within the B. subtilis MMSO and at least three promoters, one of which is sigma(beta)-dependent. The transcriptional regulation of sigA by sigma(B) provides evidence that the staphylococcal sigma(B)-dependent response is controlled at both the transcriptional and post-transcriptional level. The conservation of serp1129 across multiple gram-positive organisms and its capability to bind ATP and GTP support the need for further investigation of its role in bacterial growth.
Figures





Similar articles
-
Transcriptional characterization of the Rickettsia prowazekii major macromolecular synthesis operon.J Bacteriol. 1997 Oct;179(20):6448-52. doi: 10.1128/jb.179.20.6448-6452.1997. J Bacteriol. 1997. PMID: 9335295 Free PMC article.
-
Molecular analysis and organization of the sigmaB operon in Staphylococcus aureus.J Bacteriol. 2005 Dec;187(23):8006-19. doi: 10.1128/JB.187.23.8006-8019.2005. J Bacteriol. 2005. PMID: 16291674 Free PMC article.
-
Regulation of biofilm formation by sigma B is a common mechanism in Staphylococcus epidermidis and is not mediated by transcriptional regulation of sarA.Int J Artif Organs. 2009 Sep;32(9):584-91. doi: 10.1177/039139880903200907. Int J Artif Organs. 2009. PMID: 19856270
-
σ(ECF) factors of gram-positive bacteria: a focus on Bacillus subtilis and the CMNR group.Virulence. 2014 Jul 1;5(5):587-600. doi: 10.4161/viru.29514. Epub 2014 Jun 12. Virulence. 2014. PMID: 24921931 Free PMC article. Review.
-
STAS Domain Only Proteins in Bacterial Gene Regulation.Front Cell Infect Microbiol. 2021 Jun 21;11:679982. doi: 10.3389/fcimb.2021.679982. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34235094 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

