Molecular evolution of the three short PGRPs of the malaria vectors Anopheles gambiae and Anopheles arabiensis in East Africa
- PMID: 20067637
- PMCID: PMC2820002
- DOI: 10.1186/1471-2148-10-9
Molecular evolution of the three short PGRPs of the malaria vectors Anopheles gambiae and Anopheles arabiensis in East Africa
Abstract
Background: Immune responses to parasites, which start with pathogen recognition, play a decisive role in the control of the infection in mosquitoes. Peptidoglycan recognition proteins (PGRPs) are an important family of pattern recognition receptors that are involved in the activation of these immune reactions. Pathogen pressure can exert adaptive changes in host genes that are crucial components of the vector's defence. The aim of this study was to determine the molecular evolution of the three short PGRPs (PGRP-S1, PGRP-S2 and PGRP-S3) in the two main African malaria vectors - Anopheles gambiae and Anopheles arabiensis.
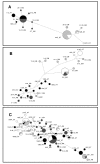
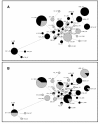
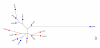
Results: Genetic diversity of An. gambiae and An. arabiensis PGRP-S1, PGRP-S2 and PGRP-S3 was investigated in samples collected from Mozambique and Tanzania. PGRP-S1 diversity was lower than for PGRP-S2 and PGRP-S3. PGRP-S1 was the only gene differentiated between the two species. All the comparisons made for PGRP-S1 showed significant P-values for Fst estimates and AMOVA confirming a clear separation between species. For PGRP-S2 and PGRP-S3 genes it was not possible to group populations either by species or by geographic region. Phylogenetic networks reinforced the results obtained by the AMOVA and Fst values. The ratio of nonsynonymous substitutions (Ka)/synonymous substitutions (Ks) for the duplicate pair PGRP-S2 and PGRP-S3 was very similar and lower than 1. The 3D model of the different proteins coded by these genes showed that amino acid substitutions were concentrated at the periphery of the protein rather than at the peptidoglycan recognition site.
Conclusions: PGRP-S1 is less diverse and showed higher divergence between An. gambiae and An. arabiensis regardless of geographic location. This probably relates to its location in the chromosome-X, while PGRP-S2 and PGRP-S3, located in chromosome-2L, showed signs of autosomal introgression. The two short PGRP genes located in the chromosome-2L were under purifying selection, which suggests functional constraints. Different types of selection acting on PGRP-S1 and PGRP-S2 and S3 might be related to their different function and catalytic activity.
Figures
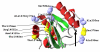
Similar articles
-
Vectorial capacity and TEP1 genotypes of Anopheles gambiae sensu lato mosquitoes on the Kenyan coast.Parasit Vectors. 2022 Dec 1;15(1):448. doi: 10.1186/s13071-022-05491-5. Parasit Vectors. 2022. PMID: 36457004 Free PMC article.
-
Patterns of selection in anti-malarial immune genes in malaria vectors: evidence for adaptive evolution in LRIM1 in Anopheles arabiensis.PLoS One. 2007 Aug 29;2(8):e793. doi: 10.1371/journal.pone.0000793. PLoS One. 2007. PMID: 17726523 Free PMC article.
-
Population genetic structure of Anopheles arabiensis and Anopheles gambiae in a malaria endemic region of southern Tanzania.Malar J. 2011 Oct 5;10:289. doi: 10.1186/1475-2875-10-289. Malar J. 2011. PMID: 21975087 Free PMC article.
-
Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories.Mol Biol Evol. 2014 Apr;31(4):889-902. doi: 10.1093/molbev/msu040. Epub 2014 Jan 9. Mol Biol Evol. 2014. PMID: 24408911 Free PMC article.
-
Population structure, speciation, and introgression in the Anopheles gambiae complex.Parassitologia. 1999 Sep;41(1-3):101-13. Parassitologia. 1999. PMID: 10697841 Review.
Cited by
-
Molecular evolution and population genetics of a Gram-negative binding protein gene in the malaria vector Anopheles gambiae (sensu lato).Parasit Vectors. 2016 Sep 23;9(1):515. doi: 10.1186/s13071-016-1800-2. Parasit Vectors. 2016. PMID: 27658383 Free PMC article.
-
Molecular evolution of a gene cluster of serine proteases expressed in the Anopheles gambiae female reproductive tract.BMC Evol Biol. 2011 Mar 19;11:72. doi: 10.1186/1471-2148-11-72. BMC Evol Biol. 2011. PMID: 21418586 Free PMC article.
-
The Negative Regulative Roles of BdPGRPs in the Imd Signaling Pathway of Bactrocera dorsalis.Cells. 2022 Jan 4;11(1):152. doi: 10.3390/cells11010152. Cells. 2022. PMID: 35011714 Free PMC article.
-
Evolution and assembly of Anopheles aquasalis's immune genes: primary malaria vector of coastal Central and South America and the Caribbean Islands.Open Biol. 2023 Jul;13(7):230061. doi: 10.1098/rsob.230061. Epub 2023 Jul 12. Open Biol. 2023. PMID: 37433331 Free PMC article.
-
Cloning, characterization and effect of TmPGRP-LE gene silencing on survival of Tenebrio molitor against Listeria monocytogenes infection.Int J Mol Sci. 2013 Nov 14;14(11):22462-82. doi: 10.3390/ijms141122462. Int J Mol Sci. 2013. PMID: 24240808 Free PMC article.
References
-
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Müller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

